Identification of P genome chromosomes in Agropyron cristatum and wheat-A. cristatum derivative lines by FISH
- PMID: 31273296
- PMCID: PMC6609639
- DOI: 10.1038/s41598-019-46197-6
Identification of P genome chromosomes in Agropyron cristatum and wheat-A. cristatum derivative lines by FISH
Abstract
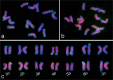
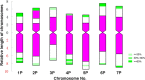
Agropyron cristatum (L.) Gaertn. (P genome) is cultivated as pasture fodder and can provide many desirable genes for wheat improvement. With the development of genomics and fluorescence in situ hybridization (FISH) technology, probes for identifying plant chromosomes were also developed. However, there are few reports on A. cristatum chromosomes. Here, FISH with the repeated sequences pAcTRT1 and pAcpCR2 enabled the identification of all diploid A. cristatum chromosomes. An integrated idiogram of A. cristatum chromosomes was constructed based on the FISH patterns of five diploid A. cristatum individuals. Structural polymorphisms of homologous chromosomes were observed not only among different individuals but also within individuals. Moreover, seventeen wheat-A. cristatum introgression lines containing different P genome chromosomes were identified with pAcTRT1 and pAcpCR2 probes. The arrangement of chromosomes in diploid A. cristatum was determined by identifying correspondence between the P chromosomes in these genetically identified introgression lines and diploid A. cristatum chromosomes. The two probes were also effective for discriminating all chromosomes of tetraploid A. cristatum, and the differences between two tetraploid A. cristatum accessions were similar to the polymorphisms among individuals of diploid A. cristatum. Collectively, the results provide an effective means for chromosome identification and phylogenetic studies of P genome chromosomes.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The launch of satellite: DNA repeats as a cytogenetic tool in discovering the chromosomal universe of wild Triticeae.Chromosoma. 2023 Jun;132(2):65-88. doi: 10.1007/s00412-023-00789-4. Epub 2023 Mar 11. Chromosoma. 2023. PMID: 36905415 Review.
-
The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes.Theor Appl Genet. 2018 Oct;131(10):2213-2227. doi: 10.1007/s00122-018-3148-9. Epub 2018 Aug 1. Theor Appl Genet. 2018. PMID: 30069594 Free PMC article.
-
Production and identification of wheat - Agropyron cristatum (1.4P) alien translocation lines.Genome. 2010 Jun;53(6):472-81. doi: 10.1139/g10-023. Genome. 2010. PMID: 20555436
-
Uncovering homeologous relationships between tetraploid Agropyron cristatum and bread wheat genomes using COS markers.Theor Appl Genet. 2019 Oct;132(10):2881-2898. doi: 10.1007/s00122-019-03394-1. Epub 2019 Jul 16. Theor Appl Genet. 2019. PMID: 31312850 Free PMC article.
-
Molecular Cytogenetic Analysis of the Introgression between Agropyron cristatum P Genome and Wheat Genome.Int J Mol Sci. 2021 Oct 18;22(20):11208. doi: 10.3390/ijms222011208. Int J Mol Sci. 2021. PMID: 34681868 Free PMC article.
Cited by
-
The Genomic Variation and Differentially Expressed Genes on the 6P Chromosomes in Wheat-Agropyron cristatum Addition Lines 5113 and II-30-5 Confer Different Desirable Traits.Int J Mol Sci. 2023 Apr 11;24(8):7056. doi: 10.3390/ijms24087056. Int J Mol Sci. 2023. PMID: 37108219 Free PMC article.
-
The launch of satellite: DNA repeats as a cytogenetic tool in discovering the chromosomal universe of wild Triticeae.Chromosoma. 2023 Jun;132(2):65-88. doi: 10.1007/s00412-023-00789-4. Epub 2023 Mar 11. Chromosoma. 2023. PMID: 36905415 Review.
-
The decreased expression of GW2 homologous genes contributed to the increased grain width and thousand‑grain weight in wheat-Dasypyrum villosum 6VS·6DL translocation lines.Theor Appl Genet. 2021 Dec;134(12):3873-3894. doi: 10.1007/s00122-021-03934-8. Epub 2021 Aug 10. Theor Appl Genet. 2021. PMID: 34374829
-
Development and application of universal ND-FISH probes for detecting P-genome chromosomes based on Agropyron cristatum transposable elements.Mol Breed. 2022 Aug 15;42(8):48. doi: 10.1007/s11032-022-01320-w. eCollection 2022 Aug. Mol Breed. 2022. PMID: 37313513 Free PMC article.
-
Draft Sequencing Crested Wheatgrass Chromosomes Identified Evolutionary Structural Changes and Genes and Facilitated the Development of SSR Markers.Int J Mol Sci. 2022 Mar 16;23(6):3191. doi: 10.3390/ijms23063191. Int J Mol Sci. 2022. PMID: 35328613 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

