Whole-Genome Sequencing of the Giant Devil Catfish, Bagarius yarrelli
- PMID: 31274158
- PMCID: PMC6681832
- DOI: 10.1093/gbe/evz143
Whole-Genome Sequencing of the Giant Devil Catfish, Bagarius yarrelli
Abstract
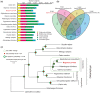
As one economically important fish in the southeastern Himalayas, the giant devil catfish (Bagarius yarrelli) has been known for its extraordinarily large body size. It can grow up to 2 m, whereas the non-Bagarius sisorids only reach 10-30 cm. Another outstanding characteristic of Bagarius species is the salmonids-like reddish flesh color. Both body size and flesh color are interesting questions in science and also valuable features in aquaculture that worth of deep investigations. Bagarius species therefore are ideal materials for studying body size evolution and color depositions in fish muscles, and also potential organisms for extensive utilization in Asian freshwater aquaculture. In a combination of Illumina and PacBio sequencing technologies, we de novo assembled a 571-Mb genome for the giant devil catfish from a total of 153.4-Gb clean reads. The scaffold and contig N50 values are 3.1 and 1.6 Mb, respectively. This genome assembly was evaluated with 93.4% of Benchmarking Universal Single-Copy Orthologs completeness, 98% of transcripts coverage, and highly homologous with a chromosome-level-based genome of channel catfish (Ictalurus punctatus). We detected that 35.26% of the genome assembly is composed of repetitive elements. Employing homology, de novo, and transcriptome-based annotations, we annotated a total of 19,027 protein-coding genes for further use. In summary, we generated the first high-quality genome assembly of the giant devil catfish, which provides an important genomic resource for its future studies such as the body size and flesh color issues, and also for facilitating the conservation and utilization of this valuable catfish.
Keywords: Bagarius yarrelli; body size; flesh color; genome assembly; giant devil catfish; whole-genome sequencing.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis.Gigascience. 2018 Nov 1;7(11):giy120. doi: 10.1093/gigascience/giy120. Gigascience. 2018. PMID: 30256939 Free PMC article.
-
Insights into Body Size Evolution: A Comparative Transcriptome Study on Three Species of Asian Sisoridae Catfish.Int J Mol Sci. 2019 Feb 21;20(4):944. doi: 10.3390/ijms20040944. Int J Mol Sci. 2019. PMID: 30795590 Free PMC article.
-
De novo transcriptome analysis of Bagarius yarrelli (Siluriformes: Sisoridae) and the search for potential SSR markers using RNA-Seq.PLoS One. 2018 Feb 9;13(2):e0190343. doi: 10.1371/journal.pone.0190343. eCollection 2018. PLoS One. 2018. PMID: 29425202 Free PMC article.
-
Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote.BMC Genomics. 2012 Nov 5;13:595. doi: 10.1186/1471-2164-13-595. BMC Genomics. 2012. PMID: 23127152 Free PMC article.
-
High-quality genome assembly of channel catfish, Ictalurus punctatus.Gigascience. 2016 Aug 22;5(1):39. doi: 10.1186/s13742-016-0142-5. Gigascience. 2016. PMID: 27549802 Free PMC article.
Cited by
-
Insights into chromosomal evolution and sex determination of Pseudobagrus ussuriensis (Bagridae, Siluriformes) based on a chromosome-level genome.DNA Res. 2022 Jun 25;29(4):dsac028. doi: 10.1093/dnares/dsac028. DNA Res. 2022. PMID: 35861402 Free PMC article.
-
Chromosome-level assembly and annotation of the blue catfish Ictalurus furcatus, an aquaculture species for hybrid catfish reproduction, epigenetics, and heterosis studies.Gigascience. 2022 Jul 9;11:giac070. doi: 10.1093/gigascience/giac070. Gigascience. 2022. PMID: 35809049 Free PMC article.
-
Telomere-to-telomere gap-free genome assembly of Euchiloglanis kishinouyei.Sci Data. 2025 May 7;12(1):757. doi: 10.1038/s41597-025-05068-8. Sci Data. 2025. PMID: 40335534 Free PMC article.
-
New complete mitogenome datasets and their characterization of the European catfish (Silurus glanis).Data Brief. 2021 Sep 23;38:107418. doi: 10.1016/j.dib.2021.107418. eCollection 2021 Oct. Data Brief. 2021. PMID: 34632016 Free PMC article.
-
Chromosome-level genome assembly of the Chinese longsnout catfish Leiocassis longirostris.Zool Res. 2021 Jul 18;42(4):417-422. doi: 10.24272/j.issn.2095-8137.2020.327. Zool Res. 2021. PMID: 34075735 Free PMC article.
References
-
- Allan JD, et al. 2005. Overfishing of inland waters. Bioscience 55(12):1041–1050.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Aparicio S, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 397:1301–1310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

