Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets
- PMID: 31274206
- PMCID: PMC6800584
- DOI: 10.1002/mp.13695
Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets
Abstract
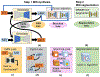
Purpose: Accurate tumor segmentation is a requirement for magnetic resonance (MR)-based radiotherapy. Lack of large expert annotated MR datasets makes training deep learning models difficult. Therefore, a cross-modality (MR-CT) deep learning segmentation approach that augments training data using pseudo MR images produced by transforming expert-segmented CT images was developed.
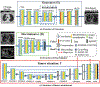
Methods: Eighty-one T2-weighted MRI scans from 28 patients with non-small cell lung cancers (nine with pretreatment and weekly MRI and the remainder with pre-treatment MRI scans) were analyzed. Cross-modality model encoding the transformation of CT to pseudo MR images resembling T2w MRI was learned as a generative adversarial deep learning network. This model was used to translate 377 expert segmented non-small cell lung cancer CT scans from the Cancer Imaging Archive into pseudo MRI that served as additional training set. This method was benchmarked against shallow learning using random forest, standard data augmentation, and three state-of-the art adversarial learning-based cross-modality data (pseudo MR) augmentation methods. Segmentation accuracy was computed using Dice similarity coefficient (DSC), Hausdorff distance metrics, and volume ratio.
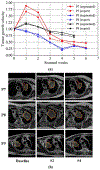
Results: The proposed approach produced the lowest statistical variability in the intensity distribution between pseudo and T2w MR images measured as Kullback-Leibler divergence of 0.069. This method produced the highest segmentation accuracy with a DSC of (0.75 ± 0.12) and the lowest Hausdorff distance of (9.36 mm ± 6.00 mm) on the test dataset using a U-Net structure. This approach produced highly similar estimations of tumor growth as an expert (P = 0.37).
Conclusions: A novel deep learning MR segmentation was developed that overcomes the limitation of learning robust models from small datasets by leveraging learned cross-modality information using a model that explicitly incorporates knowledge of tumors in modality translation to augment segmentation training. The results show the feasibility of the approach and the corresponding improvement over the state-of-the-art methods.
Keywords: cross-modality learning; data augmentation; generative adversarial networks; magnetic resonance imaging; tumor segmentation.
© 2019 American Association of Physicists in Medicine.
Figures




Similar articles
-
Deep cross-modality (MR-CT) educed distillation learning for cone beam CT lung tumor segmentation.Med Phys. 2021 Jul;48(7):3702-3713. doi: 10.1002/mp.14902. Epub 2021 May 25. Med Phys. 2021. PMID: 33905558 Free PMC article.
-
Cross-modality deep learning: Contouring of MRI data from annotated CT data only.Med Phys. 2021 Apr;48(4):1673-1684. doi: 10.1002/mp.14619. Epub 2020 Dec 13. Med Phys. 2021. PMID: 33251619 Free PMC article.
-
Self-derived organ attention for unpaired CT-MRI deep domain adaptation based MRI segmentation.Phys Med Biol. 2020 Oct 7;65(20):205001. doi: 10.1088/1361-6560/ab9fca. Phys Med Biol. 2020. PMID: 33027063
-
Deep learning based synthesis of MRI, CT and PET: Review and analysis.Med Image Anal. 2024 Feb;92:103046. doi: 10.1016/j.media.2023.103046. Epub 2023 Dec 1. Med Image Anal. 2024. PMID: 38052145 Review.
-
Deep learning techniques in PET/CT imaging: A comprehensive review from sinogram to image space.Comput Methods Programs Biomed. 2024 Jan;243:107880. doi: 10.1016/j.cmpb.2023.107880. Epub 2023 Oct 21. Comput Methods Programs Biomed. 2024. PMID: 37924769 Review.
Cited by
-
Cross2SynNet: cross-device-cross-modal synthesis of routine brain MRI sequences from CT with brain lesion.MAGMA. 2024 Apr;37(2):241-256. doi: 10.1007/s10334-023-01145-4. Epub 2024 Feb 5. MAGMA. 2024. PMID: 38315352
-
Random Multi-Channel Image Synthesis for Multiplexed Immunofluorescence Imaging.Proc Mach Learn Res. 2021 Sep;156:36-46. Proc Mach Learn Res. 2021. PMID: 34993490 Free PMC article.
-
On-board synthetic 4D MRI generation from 4D CBCT for radiotherapy of abdominal tumors: A feasibility study.Med Phys. 2024 Dec;51(12):9194-9206. doi: 10.1002/mp.17347. Epub 2024 Aug 13. Med Phys. 2024. PMID: 39137256 Free PMC article.
-
Computational approaches to detect small lesions in 18 F-FDG PET/CT scans.J Appl Clin Med Phys. 2021 Dec;22(12):125-139. doi: 10.1002/acm2.13451. Epub 2021 Oct 13. J Appl Clin Med Phys. 2021. PMID: 34643029 Free PMC article.
-
Knowledge-based radiation treatment planning: A data-driven method survey.J Appl Clin Med Phys. 2021 Aug;22(8):16-44. doi: 10.1002/acm2.13337. Epub 2021 Jul 7. J Appl Clin Med Phys. 2021. PMID: 34231970 Free PMC article. Review.
References
-
- Eisenhauer E, Therasse P, Bogaerts J et al., New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1), European journal of cancer, 2009, 45: 228–247. - PubMed
-
- Thompson RF, Valdes G, Fuller CD, Carpenter CM, Morin O, Aneja S, et al. The Future of Artificial Intelligence in Radiation Oncology. International Journal of Radiation Oncology• Biology• Physics 2018; 102(2): 247–248. - PubMed
-
- Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets, in: Advances in Neural Information Processing Systems (NIPS) 2014; p. 2672–2680.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

