PthA4AT , a 7.5-repeats transcription activator-like (TAL) effector from Xanthomonas citri ssp. citri, triggers citrus canker resistance
- PMID: 31274237
- PMCID: PMC6792138
- DOI: 10.1111/mpp.12844
PthA4AT , a 7.5-repeats transcription activator-like (TAL) effector from Xanthomonas citri ssp. citri, triggers citrus canker resistance
Abstract
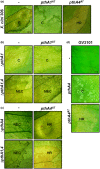
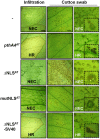
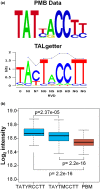
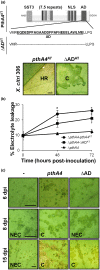
Transcription activator-like effectors (TALEs) are important effectors of Xanthomonas spp. that manipulate the transcriptome of the host plant, conferring susceptibility or resistance to bacterial infection. Xanthomonas citri ssp. citri variant AT (X. citri AT ) triggers a host-specific hypersensitive response (HR) that suppresses citrus canker development. However, the bacterial effector that elicits this process is unknown. In this study, we show that a 7.5-repeat TALE is responsible for triggering the HR. PthA4AT was identified within the pthA repertoire of X. citri AT followed by assay of the effects on different hosts. The mode of action of PthA4AT was characterized using protein-binding microarrays and testing the effects of deletion of the nuclear localization signals and activation domain on plant responses. PthA4AT is able to bind DNA and activate transcription in an effector binding element-dependent manner. Moreover, HR requires PthA4AT nuclear localization, suggesting the activation of executor resistance (R) genes in host and non-host plants. This is the first case where a TALE of unusually short length performs a biological function by means of its repeat domain, indicating that the action of these effectors to reprogramme the host transcriptome following nuclear localization is not limited to 'classical' TALEs.
Keywords: Nicotiana benthamiana; Xanthomonas citri; citrus; hypersensitive response (HR); transcription activator-like (TAL) effectors.
© 2019 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures



References
-
- Al‐Saadi, A. , Reddy, J.D. , Duan, Y.P. , Brunings, A.M. , Yuan, Q. and Gabriel, D.W. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Mol. Plant‐Microbe Interact. 20, 934–943. - PubMed
-
- Behlau, F. and Belasque, J. , Graham, J.H. and Leite, R.P. (2010) Effect of frequency of copper applications on control of citrus canker and the yield of young bearing sweet orange trees. Crop Prot. 29, 300–305.
-
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. - PubMed
-
- Boch, J. , Bonas, U. and Lahaye, T. (2014) TAL effectors‐pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

