Effect of Single Dose of Antimicrobial Administration at Birth on Fecal Microbiota Development and Prevalence of Antimicrobial Resistance Genes in Piglets
- PMID: 31275295
- PMCID: PMC6593251
- DOI: 10.3389/fmicb.2019.01414
Effect of Single Dose of Antimicrobial Administration at Birth on Fecal Microbiota Development and Prevalence of Antimicrobial Resistance Genes in Piglets
Abstract
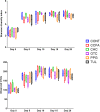
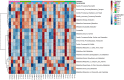
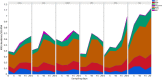
Optimization of antimicrobial use in swine management systems requires full understanding of antimicrobial-induced changes on the developmental dynamics of gut microbiota and the prevalence of antimicrobial resistance genes (ARGs). The purpose of this study was to evaluate the impacts of early life antimicrobial intervention on fecal microbiota development, and prevalence of selected ARGs (ermB, tetO, tetW, tetC, sulI, sulII, and blaC TX-M) in neonatal piglets. A total of 48 litters were randomly allocated into one of six treatment groups soon after birth. Treatments were as follows: control (CONT), ceftiofur crystalline free acid (CCFA), ceftiofur hydrochloride (CHC), oxytetracycline (OTC), procaine penicillin G (PPG), and tulathromycin (TUL). Fecal swabs were collected from piglets at days 0 (prior to treatment), 5, 10, 15, and 20 post treatment. Sequencing analysis of the V3-V4 hypervariable region of the 16S rRNA gene and selected ARGs were performed using the Illumina Miseq platform. Our results showed that, while early life antimicrobial prophylaxis had no effect on individual weight gain, or mortality, it was associated with minor shifts in the composition of fecal microbiota and noticeable changes in the abundance of selected ARGs. Unifrac distance metrics revealed that the microbial communities of the piglets that received different treatments (CCFA, CHC, OTC, PPG, and TUL) did not cluster distinctly from CONT piglets. Compared to CONT group, PPG-treated piglets exhibited a significant increase in the relative abundance of ermB and tetW at day 20 of life. Tulathromycin treatment also resulted in a significant increase in the abundance of tetW at days 10 and 20, and ermB at day 20. Collectively, these results demonstrate that the shifts in fecal microbiota structure caused by perinatal antimicrobial intervention are modest and limited to particular groups of microbial taxa. However, early life PPG and TUL intervention could promote the selection of ARGs in herds. While additional investigations are required to explore the consistency of these findings across larger populations, these results could open the door to new perspectives on the utility of early life antimicrobial administration to healthy neonates in swine management systems.
Keywords: antimicrobial; fecal; microbiota; neonatal piglets; resistance genes.
Figures





Similar articles
-
Metagenomic Analysis of the Fecal Archaeome in Suckling Piglets Following Perinatal Tulathromycin Metaphylaxis.Animals (Basel). 2021 Jun 18;11(6):1825. doi: 10.3390/ani11061825. Animals (Basel). 2021. PMID: 34207278 Free PMC article.
-
Negligible Impact of Perinatal Tulathromycin Metaphylaxis on the Developmental Dynamics of Fecal Microbiota and Their Accompanying Antimicrobial Resistome in Piglets.Front Microbiol. 2019 Apr 5;10:726. doi: 10.3389/fmicb.2019.00726. eCollection 2019. Front Microbiol. 2019. PMID: 31024502 Free PMC article.
-
Impact of parenteral antimicrobial administration on the structure and diversity of the fecal microbiota of growing pigs.Microb Pathog. 2018 May;118:220-229. doi: 10.1016/j.micpath.2018.03.035. Epub 2018 Mar 22. Microb Pathog. 2018. PMID: 29578067
-
Microbial shifts in the swine nasal microbiota in response to parenteral antimicrobial administration.Microb Pathog. 2018 Aug;121:210-217. doi: 10.1016/j.micpath.2018.05.028. Epub 2018 May 24. Microb Pathog. 2018. PMID: 29803848
-
Antimicrobial Effects on Swine Gastrointestinal Microbiota and Their Accompanying Antibiotic Resistome.Front Microbiol. 2019 May 15;10:1035. doi: 10.3389/fmicb.2019.01035. eCollection 2019. Front Microbiol. 2019. PMID: 31156580 Free PMC article. Review.
Cited by
-
Replacing dietary antibiotics with 0.20% l-glutamine in swine nursery diets: impact on intestinal physiology and the microbiome following weaning and transport.J Anim Sci. 2021 Jun 1;99(6):skab091. doi: 10.1093/jas/skab091. J Anim Sci. 2021. PMID: 33755169 Free PMC article.
-
Insights on the effects of antimicrobial and heavy metal usage on the antimicrobial resistance profiles of pigs based on culture-independent studies.Vet Res. 2023 Feb 23;54(1):14. doi: 10.1186/s13567-023-01143-3. Vet Res. 2023. PMID: 36823539 Free PMC article. Review.
-
Cytotoxicity and degradation product identification of thermally treated ceftiofur.RSC Adv. 2020 May 13;10(31):18407-18417. doi: 10.1039/c9ra10289b. eCollection 2020 May 10. RSC Adv. 2020. PMID: 35517214 Free PMC article.
-
Metagenomic Analysis of the Fecal Archaeome in Suckling Piglets Following Perinatal Tulathromycin Metaphylaxis.Animals (Basel). 2021 Jun 18;11(6):1825. doi: 10.3390/ani11061825. Animals (Basel). 2021. PMID: 34207278 Free PMC article.
-
Impact of Parenteral Ceftiofur on Developmental Dynamics of Early Life Fecal Microbiota and Antibiotic Resistome in Neonatal Lambs.Antibiotics (Basel). 2025 Apr 25;14(5):434. doi: 10.3390/antibiotics14050434. Antibiotics (Basel). 2025. PMID: 40426501 Free PMC article.
References
-
- Bian G., Ma S., Zhu Z., Su Y., Zoetendal E. G., Mackie R., et al. (2016). Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ. Microbiol. 18 1566–1577. 10.1111/1462-2920.13272 - DOI - PubMed
-
- Bischoff K. M., White D. G., McDermott P. F., Zhao S., Gaines S., Maurer J. J., et al. (2002). Characterization of chloramphenicol resistance in beta-hemolytic Escherichia coli associated with diarrhea in neonatal swine. J. Clin. Microbiol. 40 389–394. 10.1128/JCM.40.2.389-394.2002 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

