A Modular Cytokine Analysis Method Reveals Novel Associations With Clinical Phenotypes and Identifies Sets of Co-signaling Cytokines Across Influenza Natural Infection Cohorts and Healthy Controls
- PMID: 31275311
- PMCID: PMC6594355
- DOI: 10.3389/fimmu.2019.01338
A Modular Cytokine Analysis Method Reveals Novel Associations With Clinical Phenotypes and Identifies Sets of Co-signaling Cytokines Across Influenza Natural Infection Cohorts and Healthy Controls
Abstract
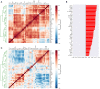
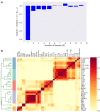
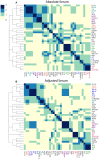
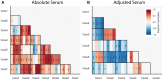
Cytokines and chemokines are key signaling molecules of the immune system. Recent technological advances enable measurement of multiplexed cytokine profiles in biological samples. These profiles can then be used to identify potential biomarkers of a variety of clinical phenotypes. However, testing for such associations for each cytokine separately ignores the highly context-dependent covariation in cytokine secretion and decreases statistical power to detect associations due to multiple hypothesis testing. Here we present CytoMod-a novel data-driven approach for analysis of cytokine profiles that uses unsupervised clustering and regression to identify putative functional modules of co-signaling cytokines. Each module represents a biosignature of co-signaling cytokines. We applied this approach to three independent clinical cohorts of subjects naturally infected with influenza in which cytokine profiles and clinical phenotypes were collected. We found that in two out of three cohorts, cytokine modules were significantly associated with clinical phenotypes, and in many cases these associations were stronger than the associations of the individual cytokines within them. By comparing cytokine modules across datasets, we identified cytokine "cores"-specific subsets of co-expressed cytokines that clustered together across the three cohorts. Cytokine cores were also associated with clinical phenotypes. Interestingly, most of these cores were also co-expressed in a cohort of healthy controls, suggesting that in part, patterns of cytokine co-signaling may be generalizable. CytoMod can be readily applied to any cytokine profile dataset regardless of measurement technology, increases the statistical power to detect associations with clinical phenotypes and may help shed light on the complex co-signaling networks of cytokines in both health and infection.
Keywords: biomarker; chemokines; cytokines; influenza; innate immunology.
Figures




References
-
- Miller MD, Krangel MS. Biology and biochemistry of the chemokines: a family of chemotactic and inflammatory cytokines. Crit Rev Immunol. (1992) 12:17–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

