Gauging genetic diversity of generalists: A test of genetic and ecological generalism with RNA virus experimental evolution
- PMID: 31275611
- PMCID: PMC6599687
- DOI: 10.1093/ve/vez019
Gauging genetic diversity of generalists: A test of genetic and ecological generalism with RNA virus experimental evolution
Abstract
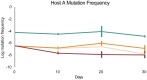
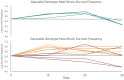
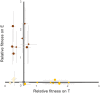
Generalist viruses, those with a comparatively larger host range, are considered more likely to emerge on new hosts. The potential to emerge in new hosts has been linked to viral genetic diversity, a measure of evolvability. However, there is no consensus on whether infecting a larger number of hosts leads to higher genetic diversity, or whether diversity is better maintained in a homogeneous environment, similar to the lifestyle of a specialist virus. Using experimental evolution with the RNA bacteriophage phi6, we directly tested whether genetic generalism (carrying an expanded host range mutation) or environmental generalism (growing on heterogeneous hosts) leads to viral populations with more genetic variation. Sixteen evolved viral lineages were deep sequenced to provide genetic evidence for population diversity. When evolved on a single host, specialist and generalist genotypes both maintained the same level of diversity (measured by the number of single nucleotide polymorphisms (SNPs) above 1%, P = 0.81). However, the generalist genotype evolved on a single host had higher SNP levels than generalist lineages under two heterogeneous host passaging schemes (P = 0.001, P < 0.001). RNA viruses' response to selection in alternating hosts reduces standing genetic diversity compared to those evolving in a single host to which the virus is already well-adapted.
Keywords: dsRNA virus; generalist; genetic diversity; phage evolution.
Figures


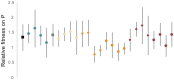
Similar articles
-
Variation in within-host replication kinetics among virus genotypes provides evidence of specialist and generalist infection strategies across three salmonid host species.Virus Evol. 2022 Aug 24;8(2):veac079. doi: 10.1093/ve/veac079. eCollection 2022. Virus Evol. 2022. PMID: 36101884 Free PMC article.
-
Evolution in spatially mixed host environments increases divergence for evolved fitness and intrapopulation genetic diversity in RNA viruses.Virus Evol. 2016 Jan 20;2(1):vev022. doi: 10.1093/ve/vev022. eCollection 2016 Jan. Virus Evol. 2016. PMID: 27774292 Free PMC article.
-
Local adaptation of plant viruses: lessons from experimental evolution.Mol Ecol. 2017 Apr;26(7):1711-1719. doi: 10.1111/mec.13836. Epub 2016 Sep 22. Mol Ecol. 2017. PMID: 27612225 Review.
-
Defects in plant immunity modulate the rates and patterns of RNA virus evolution.Virus Evol. 2022 Jun 20;8(2):veac059. doi: 10.1093/ve/veac059. eCollection 2022. Virus Evol. 2022. PMID: 35821716 Free PMC article.
-
Evolution of pathogen virulence: the role of variation in host phenotype.Proc Biol Sci. 2001 Apr 7;268(1468):755-60. doi: 10.1098/rspb.2000.1582. Proc Biol Sci. 2001. PMID: 11321065 Free PMC article. Review.
Cited by
-
Experimental Evolution Studies in Φ6 Cystovirus.Viruses. 2024 Jun 18;16(6):977. doi: 10.3390/v16060977. Viruses. 2024. PMID: 38932268 Free PMC article. Review.
-
Virus Prevalence and Genetic Diversity Across a Wild Bumblebee Community.Front Microbiol. 2021 Apr 22;12:650747. doi: 10.3389/fmicb.2021.650747. eCollection 2021. Front Microbiol. 2021. PMID: 33967987 Free PMC article.
-
Contrasting patterns of genetic and phenotypic divergence of two sympatric congeners, Phragmites australis and P. hirsuta, in heterogeneous habitats.Front Plant Sci. 2023 Dec 12;14:1299128. doi: 10.3389/fpls.2023.1299128. eCollection 2023. Front Plant Sci. 2023. PMID: 38162310 Free PMC article.
-
Remarkably coherent population structure for a dominant Antarctic Chlorobium species.Microbiome. 2021 Nov 26;9(1):231. doi: 10.1186/s40168-021-01173-z. Microbiome. 2021. PMID: 34823595 Free PMC article.
References
-
- Aguirre J., Lázaro E., Manrubia S. C. (2009) ‘A Trade-off between Neutrality and Adaptability Limits the Optimization of Viral Quasispecies’, Journal of Theoretical Biology, 261: 148–55. - PubMed
-
- Ali J. G., Agrawal A. A. (2012) ‘Specialist Versus Generalist Insect Herbivores and Plant Defense’, Trends in Plant Science, 17: 293–302. - PubMed
-
- Alto B. W. et al. (2013) ‘Stochastic Temperatures Impede RNA Virus Adaptation’, Evolution, 67: 969–79. - PubMed
LinkOut - more resources
Full Text Sources

