Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish
- PMID: 31277329
- PMCID: PMC6789471
- DOI: 10.3390/vaccines7030060
Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish
Abstract
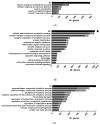
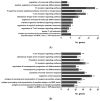
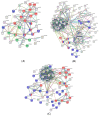
In recent years, fish nucleated red blood cells (RBCs) have been implicated in the response against viral infections. We have demonstrated that rainbow trout RBCs can express the antigen encoded by a DNA vaccine against viral hemorrhagic septicemia virus (VHSV) and mount an immune response to the antigen in vitro. In this manuscript, we show, for the first time, the role of RBCs in the immune response triggered by DNA immunization of rainbow trout with glycoprotein G of VHSV (GVHSV). Transcriptomic and proteomic profiles of RBCs revealed genes and proteins involved in antigen processing and presentation of exogenous peptide antigen via MHC class I, the Fc receptor signaling pathway, the autophagy pathway, and the activation of the innate immune response, among others. On the other hand, GVHSV-transfected RBCs induce specific antibodies against VHSV in the serum of rainbow trout which shows that RBCs expressing a DNA vaccine are able to elicit a humoral response. These results open a new direction in the research of vaccination strategies for fish since rainbow trout RBCs actively participate in the innate and adaptive immune response in DNA vaccination. Based on our findings, we suggest the use of RBCs as target cells or carriers for the future design of novel vaccine strategies.
Keywords: GVHSV; antigen presentation; erythrocytes; proteome; rainbow trout; red blood cells; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest
Figures


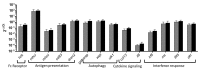

Similar articles
-
Rainbow Trout Erythrocytes ex vivo Transfection With a DNA Vaccine Encoding VHSV Glycoprotein G Induces an Antiviral Immune Response.Front Immunol. 2018 Oct 29;9:2477. doi: 10.3389/fimmu.2018.02477. eCollection 2018. Front Immunol. 2018. PMID: 30429850 Free PMC article.
-
Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers.Cells. 2019 Apr 27;8(5):386. doi: 10.3390/cells8050386. Cells. 2019. PMID: 31035565 Free PMC article.
-
Integrated Transcriptomic and Proteomic Analysis of Red Blood Cells from Rainbow Trout Challenged with VHSV Point Towards Novel Immunomodulant Targets.Vaccines (Basel). 2019 Jul 9;7(3):63. doi: 10.3390/vaccines7030063. Vaccines (Basel). 2019. PMID: 31324030 Free PMC article.
-
IFIT5 Participates in the Antiviral Mechanisms of Rainbow Trout Red Blood Cells.Front Immunol. 2019 Apr 16;10:613. doi: 10.3389/fimmu.2019.00613. eCollection 2019. Front Immunol. 2019. PMID: 31040842 Free PMC article.
-
Identification of diverse defense mechanisms in rainbow trout red blood cells in response to halted replication of VHS virus.F1000Res. 2017 Nov 6;6:1958. doi: 10.12688/f1000research.12985.2. eCollection 2017. F1000Res. 2017. PMID: 29527292 Free PMC article.
Cited by
-
An SNP-Based Genetic Map and QTL Mapping for Growth Traits in the Red-Spotted Grouper (Epinephelus akaara).Genes (Basel). 2019 Oct 12;10(10):793. doi: 10.3390/genes10100793. Genes (Basel). 2019. PMID: 31614822 Free PMC article.
-
Innate immune response of rainbow trout erythrocytes to spinycterins expressing a downsized viral fragment of viral haemorrhagic septicaemia virus.Aquaculture. 2023 Apr 15;568:739303. doi: 10.1016/j.aquaculture.2023.739303. Aquaculture. 2023. PMID: 38533126 Free PMC article.
-
Viral vaccines promote endoplasmic reticulum stress-induced unfolding protein response in teleost erythrocytes.Eur J Cell Biol. 2025 Jun;104(2):151490. doi: 10.1016/j.ejcb.2025.151490. Epub 2025 Apr 9. Eur J Cell Biol. 2025. PMID: 40252498 Free PMC article.
-
Tilapia Lake Virus Vaccine Development: A Review on the Recent Advances.Vaccines (Basel). 2023 Jan 23;11(2):251. doi: 10.3390/vaccines11020251. Vaccines (Basel). 2023. PMID: 36851129 Free PMC article. Review.
-
Single-Cell RNA Sequencing Reveals Microevolution of the Stickleback Immune System.Genome Biol Evol. 2023 Apr 6;15(4):evad053. doi: 10.1093/gbe/evad053. Genome Biol Evol. 2023. PMID: 37039516 Free PMC article.
References
-
- Food and Agriculture Organization of the United Nations (FAO) The State Of World Fisheries and Aquaculture. Meeting the Sustainable Development Goals. FAO; Rome, Italy: 2018.
-
- Kurath G. Fish. Novirhabdoviruses. Caister Academic Press; Norfolk, UK: 2012.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

