Expanding the Diversity of Myoviridae Phages Infecting Lactobacillus plantarum-A Novel Lineage of Lactobacillus Phages Comprising Five New Members
- PMID: 31277436
- PMCID: PMC6669764
- DOI: 10.3390/v11070611
Expanding the Diversity of Myoviridae Phages Infecting Lactobacillus plantarum-A Novel Lineage of Lactobacillus Phages Comprising Five New Members
Abstract
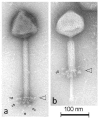
Lactobacillus plantarum is a bacterium with probiotic properties and promising applications in the food industry and agriculture. So far, bacteriophages of this bacterium have been moderately addressed. We examined the diversity of five new L. plantarum phages via whole genome shotgun sequencing and in silico protein predictions. Moreover, we looked into their phylogeny and their potential genomic similarities to other complete phage genome records through extensive nucleotide and protein comparisons. These analyses revealed a high degree of similarity among the five phages, which extended to the vast majority of predicted virion-associated proteins. Based on these, we selected one of the phages as a representative and performed transmission electron microscopy and structural protein sequencing tests. Overall, the results suggested that the five phages belong to the family Myoviridae, they have a long genome of 137,973-141,344 bp, a G/C content of 36.3-36.6% that is quite distinct from their host's, and surprisingly, 7 to 15 tRNAs. Only an average 41/174 of their predicted genes were assigned a function. The comparative analyses unraveled considerable genetic diversity for the five L. plantarum phages in this study. Hence, the new genus "Semelevirus" was proposed, comprising exclusively of the five phages. This novel lineage of Lactobacillus phages provides further insight into the genetic heterogeneity of phages infecting Lactobacillus sp. The five new Lactobacillus phages have potential value for the development of more robust starters through, for example, the selection of mutants insensitive to phage infections. The five phages could also form part of phage cocktails, which producers would apply in different stages of L. plantarum fermentations in order to create a range of organoleptic outputs.
Keywords: Lactobacillus plantarum; annotation; comparative genomics; diversity; isolation; new genus; phage; phylogenetics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- König H., Fröhlich J. Lactic Acid Bacteria. In: König H., Unden G., Fröhlich J., editors. Biology of Microorganisms on Grapes, in Must and in Wine. Springer International Publishing; Cham, Switzerland: 2017. pp. 3–41.
-
- Lamont J.R., Wilkins O., Bywater-Ekegärd M., Smith D.L. From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 2017;111:1–9. doi: 10.1016/j.soilbio.2017.03.015. - DOI
-
- Du Toit M., Engelbrecht L., Lerm E., Krieger-Weber S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures—An Overview. Food Bioprocess Technol. 2011;4:876–906. doi: 10.1007/s11947-010-0448-8. - DOI
-
- Lerm E., Engelbrecht L., Du Toit M. Selection and characterisation of Oenococcus oeni and Lactobacillus plantarum South African wine isolates for use as malolactic fermentation starter cultures. S. Afr. J. Enol. Vitic. 2011;32:280–295. doi: 10.21548/32-2-1388. - DOI
-
- López I., López R., Santamaría P., Torres C., Ruiz-Larrea F. Performance of malolactic fermentation by inoculation of selected Lactobacillus plantarum and Oenococcus oeni strains isolated from Rioja red wines. Vitis. 2008;47:123–129.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

