Novel significant stage-specific differentially expressed genes in hepatocellular carcinoma
- PMID: 31277598
- PMCID: PMC6612102
- DOI: 10.1186/s12885-019-5838-3
Novel significant stage-specific differentially expressed genes in hepatocellular carcinoma
Abstract
Background: Liver cancer is among top deadly cancers worldwide with a very poor prognosis, and the liver is a vulnerable site for metastases of other cancers. Early diagnosis is crucial for treatment of the predominant liver cancers, namely hepatocellular carcinoma (HCC). Here we developed a novel computational framework for the stage-specific analysis of HCC.
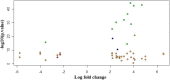
Methods: Using publicly available clinical and RNA-Seq data of cancer samples and controls and the AJCC staging system, we performed a linear modelling analysis of gene expression across all stages and found significant genome-wide changes in the log fold-change of gene expression in cancer samples relative to control. To identify genes that were stage-specific controlling for confounding differential expression in other stages, we developed a set of six pairwise contrasts between the stages and enforced a p-value threshold (< 0.05) for each such contrast. Genes were specific for a stage if they passed all the significance filters for that stage. The monotonicity of gene expression with cancer progression was analyzed with a linear model using the cancer stage as a numeric variable.
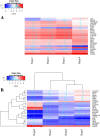
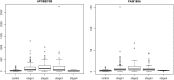
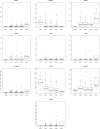
Results: Our analysis yielded two stage-I specific genes (CA9, WNT7B), two stage-II specific genes (APOBEC3B, FAM186A), ten stage-III specific genes including DLG5, PARI, NCAPG2, GNMT and XRCC2, and 35 stage-IV specific genes including GABRD, PGAM2, PECAM1 and CXCR2P1. Overexpression of DLG5 was found to be tumor-promoting contrary to the cancer literature on this gene. Further, GABRD was found to be signifincantly monotonically upregulated across stages. Our work has revealed 1977 genes with significant monotonic patterns of expression across cancer stages. NDUFA4L2, CRHBP and PIGU were top genes with monotonic changes of expression across cancer stages that could represent promising targets for therapy. Comparison with gene signatures from the BCLC staging system identified two genes, HSP90AB1 and ARHGAP42. Gene set enrichment analysis indicated overrepresented pathways specific to each stage, notably viral infection pathways in HCC initiation.
Conclusions: Our study identified novel significant stage-specific differentially expressed genes which could enhance our understanding of the molecular determinants of hepatocellular carcinoma progression. Our findings could serve as biomarkers that potentially underpin diagnosis as well as pinpoint therapeutic targets.
Keywords: Cancer progression; Differentially expressed genes; HCC stages; LIHC transcriptomics; Linear modelling; Metastasis; Monotonic expression; Pairwise contrasts; Significance analysis; Stage-specific biomarkers; Tumorigenesis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures










References
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jäger N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdés-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. - DOI - PMC - PubMed
-
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC Cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–99. doi: 10.3322/caac.21388. - DOI - PubMed
-
- Arensman MD, Kovochich AN, Kulikauskas RM, Lay AR, Yang PT, Li X, Donahue T, Major MB, Moon RT, Chien AJ, Dawson DW. WNT7B mediates autocrine WNT/β-catenin signaling and anchorage-independent growth in pancreatic adenocarcinoma. Oncogene. 2014;33(7):899. doi: 10.1038/onc.2013.23. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

