Leveraging -omics for asthma endotyping
- PMID: 31277743
- PMCID: PMC6721613
- DOI: 10.1016/j.jaci.2019.05.015
Leveraging -omics for asthma endotyping
Abstract
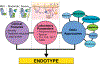
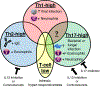
Asthma is a highly heterogeneous disease, often manifesting with wheeze, dyspnea, chest tightness, and cough as prominent symptoms. The eliciting factors, natural history, underlying molecular biology, and clinical management of asthma vary highly among affected subjects. Because of this variation, many efforts have gone into subtyping asthma. Endotypes are subtypes of disease based on distinct pathophysiologic mechanisms. Endotypes can be clinically useful because they organize our mechanistic understanding of heterogeneous diseases and can direct treatment toward modalities that are likely to be the most effective. Asthma endotyping can be shaped by clinical features, laboratory parameters, and/or -omics approaches. We discuss the application of -omics approaches, including transcriptomics, epigenomics, microbiomics, metabolomics, and proteomics, to asthma endotyping. -Omics approaches have provided supporting evidence for many existing endotyping paradigms and also suggested novel ways to conceptualize asthma endotypes. Although endotypes based on single -omics approaches are relatively common, their integrated multi-omics application to asthma endotyping has been more limited thus far. We discuss paths forward to integrate multi-omics with clinical features and laboratory parameters to achieve the goal of precise asthma endotypes.
Keywords: -ome; -omic; Asthma; cluster; endotype; epigenome; integrate; metabolome; microbiome; multi-ome; phenotype; proteome; transcriptome.
Copyright © 2019 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures


References
-
- Casale TB. Biologics and biomarkers for asthma, urticaria, and nasal polyposis. J Allergy Clin Immunol 2017; 139:1411–21. - PubMed
-
- Skloot GS. Asthma phenotypes and endotypes: a personalized approach to treatment. Curr Opin Pulm Med 2016; 22:3–9. - PubMed
-
- Nagasaki T, Matsumoto H, Izuhara K, Kanemitsu Y, Tohda Y, Horiguchi T, et al. Utility of serum periostin in combination with exhaled nitric oxide in the management of asthma. Allergology International 2017; 66:404–10. - PubMed

