A novel motif in the proximal C-terminus of Pannexin 1 regulates cell surface localization
- PMID: 31278290
- PMCID: PMC6611761
- DOI: 10.1038/s41598-019-46144-5
A novel motif in the proximal C-terminus of Pannexin 1 regulates cell surface localization
Abstract
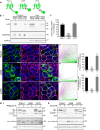
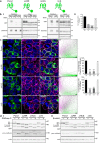
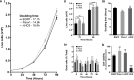
The Pannexin 1 (Panx1) ion and metabolite channel is expressed in a wide variety of cells where it regulates a number of cell behaviours including proliferation and differentiation. Panx1 is expressed on the cell surface as well as intracellular membranes. Previous work suggests that a region within the proximal Panx1 C-terminus (Panx1CT) regulates cell surface localization. Here we report the discovery of a putative leucine-rich repeat (LRR) motif in the proximal Panx1CT necessary for Panx1 cell surface expression in HEK293T cells. Deletion of the putative LRR motif results in significant loss of Panx1 cell surface distribution. Outcomes of complementary cell surface oligomerization and glycosylation state analyses were consistent with reduced cell surface expression of Panx1 LRR deletion mutants. Of note, the oligomerization analysis revealed the presence of putative dimers and trimers of Panx1 at the cell surface. Expression of Panx1 increased HEK293T cell growth and reduced doubling time, while expression of a Panx1 LRR deletion mutant (highly conserved segment) did not reproduce this effect. In summary, here we discovered the presence of a putative LRR motif in the Panx1CT that impacts on Panx1 cell surface localization. Overall these findings provide new insights into the molecular mechanisms underlying C-terminal regulation of Panx1 trafficking and raise potential new lines of investigation with respect to Panx1 oligomerization and glycosylation.
Conflict of interest statement
The authors declare no competing interests. A disclosure has been made with the University of Victoria Research Partnerships and Knowledge Mobilization, and a provisional patent application has been filed for a peptide targeting a Panx1-Crmp2 interaction (US Application No. 62/767,806) that does not relate directly to this study.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

