Delayed recruiting of TPD52 to lipid droplets - evidence for a "second wave" of lipid droplet-associated proteins that respond to altered lipid storage induced by Brefeldin A treatment
- PMID: 31278300
- PMCID: PMC6611826
- DOI: 10.1038/s41598-019-46156-1
Delayed recruiting of TPD52 to lipid droplets - evidence for a "second wave" of lipid droplet-associated proteins that respond to altered lipid storage induced by Brefeldin A treatment
Abstract
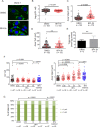
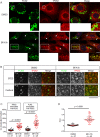
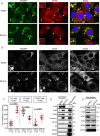
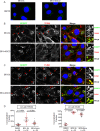
Tumor protein D52 (TPD52) is amplified and overexpressed in breast and prostate cancers which are frequently characterised by dysregulated lipid storage and metabolism. TPD52 expression increases lipid storage in mouse 3T3 fibroblasts, and co-distributes with the Golgi marker GM130 and lipid droplets (LDs). We examined the effects of Brefeldin A (BFA), a fungal metabolite known to disrupt the Golgi structure, in TPD52-expressing 3T3 cells, and in human AU565 and HMC-1-8 breast cancer cells that endogenously express TPD52. Five-hour BFA treatment reduced median LD numbers, but increased LD sizes. TPD52 knockdown decreased both LD sizes and numbers, and blunted BFA's effects on LD numbers. Following BFA treatment for 1-3 hours, TPD52 co-localised with the trans-Golgi network protein syntaxin 6, but after 5 hours BFA treatment, TPD52 showed increased co-localisation with LDs, which was disrupted by microtubule depolymerising agent nocodazole. BFA treatment also increased perilipin (PLIN) family protein PLIN3 but reduced PLIN2 detection at LDs in TPD52-expressing 3T3 cells, with PLIN3 recruitment to LDs preceding that of TPD52. An N-terminally deleted HA-TPD52 mutant (residues 40-184) almost exclusively targeted to LDs in both vehicle and BFA treated cells. In summary, delayed recruitment of TPD52 to LDs suggests that TPD52 participates in a temporal hierarchy of LD-associated proteins that responds to altered LD packaging requirements induced by BFA treatment.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

