Application of Proteomics Profiling for Biomarker Discovery in Hypertrophic Cardiomyopathy
- PMID: 31278493
- PMCID: PMC7102897
- DOI: 10.1007/s12265-019-09896-z
Application of Proteomics Profiling for Biomarker Discovery in Hypertrophic Cardiomyopathy
Abstract
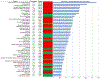
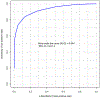
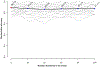
High-throughput proteomics profiling has never been applied to discover biomarkers in patients with hypertrophic cardiomyopathy (HCM). The objective was to identify plasma protein biomarkers that can distinguish HCM from controls. We performed a case-control study of patients with HCM (n = 15) and controls (n = 22). We carried out plasma proteomics profiling of 1129 proteins using the SOMAscan assay. We used the sparse partial least squares discriminant analysis to identify 50 most discriminant proteins. We also determined the area under the curve (AUC) of the receiver operating characteristic curve using the Monte Carlo cross validation with balanced subsampling. The average AUC was 0.94 (95% confidence interval, 0.82-1.00) and the discriminative accuracy was 89%. In HCM, 13 out of the 50 proteins correlated with troponin I and 12 with New York Heart Association class. Proteomics profiling can be used to elucidate protein biomarkers that distinguish HCM from controls.
Keywords: Biomarker discovery; Hypertrophic cardiomyopathy; Proteomics.
Conflict of interest statement
Figures

References
-
- Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, & Bild DE (1995). Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA study. Coronary Artery Risk Development in (Young) Adults. Circulation, 92, 785–789 - PubMed
-
- Kiddle SJ, Sattlecker M, Proitsi P, Simmons A, Westman E, Bazenet C, et al. (2014). Candidate blood proteome markers of Alzheimer’s disease onset and progression: a systematic review and replication study. Journal of Alzheimer’s Disease, 38, 515–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

