Delineating the Plausible Molecular Vaccine Candidates and Drug Targets of Multidrug-Resistant Acinetobacter baumannii
- PMID: 31281799
- PMCID: PMC6596342
- DOI: 10.3389/fcimb.2019.00203
Delineating the Plausible Molecular Vaccine Candidates and Drug Targets of Multidrug-Resistant Acinetobacter baumannii
Abstract
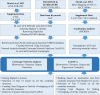
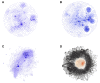
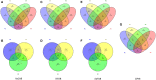
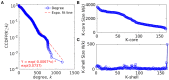
Nosocomial infections have become alarming with the increase of multidrug-resistant bacterial strains of Acinetobacter baumannii. Being the causative agent in ~80% of the cases, these pathogenic gram-negative species could be deadly for hospitalized patients, especially in intensive care units utilizing ventilators, urinary catheters, and nasogastric tubes. Primarily infecting an immuno-compromised system, they are resistant to most antibiotics and are the root cause of various types of opportunistic infections including but not limited to septicemia, endocarditis, meningitis, pneumonia, skin, and wound sepsis and even urinary tract infections. Conventional experimental methods including typing, computational methods encompassing comparative genomics, and combined methods of reverse vaccinology and proteomics had been proposed to differentiate and develop vaccines and/or drugs for several outbreak strains. However, identifying proteins suitable enough to be posed as drug targets and/or molecular vaccines against the multidrug-resistant pathogenic bacterial strains has probably remained an open issue to address. In these cases of novel protein identification, the targets either are uncharacterized or have been unable to confer the most coveted protection either in the form of molecular vaccine candidates or as drug targets. Here, we report a strategic approach with the 3,766 proteins from the whole genome of A. baumannii ATCC19606 (AB) to rationally identify plausible candidates and propose them as future molecular vaccine candidates and/or drug targets. Essentially, we started with mapping the vaccine candidates (VaC) and virulence factors (ViF) of A. baumannii strain AYE onto strain ATCC19606 to identify them in the latter. We move on to build small networks of VaC and ViF to conceptualize their position in the network space of the whole genomic protein interactome (GPIN) and rationalize their candidature for drugs and/or molecular vaccines. To this end, we propose new sets of known proteins unearthed from interactome built using key factors, KeF, potent enough to compete with VaC and ViF. Our method is the first of its kind to propose, albeit theoretically, a rational approach to identify crucial proteins and pose them for candidates of vaccines and/or drugs effective enough to combat the deadly pathogenic threats of A. baumannii.
Keywords: Acinetobacter baumannii; drug targets; network analysis; nosocomial infection; vaccine candidates.
Figures
Similar articles
-
Promising Acinetobacter baumannii Vaccine Candidates and Drug Targets in Recent Years.Front Immunol. 2022 May 26;13:900509. doi: 10.3389/fimmu.2022.900509. eCollection 2022. Front Immunol. 2022. PMID: 35720310 Free PMC article. Review.
-
Reverse Vaccinology Approach to Potential Vaccine Candidates Against Acinetobacter baumannii.Methods Mol Biol. 2019;1946:329-336. doi: 10.1007/978-1-4939-9118-1_29. Methods Mol Biol. 2019. PMID: 30798567 Review.
-
Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii.PLoS One. 2013 Oct 8;8(10):e77631. doi: 10.1371/journal.pone.0077631. eCollection 2013. PLoS One. 2013. PMID: 24116234 Free PMC article.
-
Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology.Int J Mol Sci. 2017 Feb 21;18(2):458. doi: 10.3390/ijms18020458. Int J Mol Sci. 2017. PMID: 28230771 Free PMC article.
-
In Silico Identification of Probable Drug and Vaccine Candidates Against Antibiotic-Resistant Acinetobacter baumannii.Microb Drug Resist. 2020 May;26(5):456-467. doi: 10.1089/mdr.2019.0236. Epub 2019 Nov 19. Microb Drug Resist. 2020. PMID: 31742478
Cited by
-
Promising Acinetobacter baumannii Vaccine Candidates and Drug Targets in Recent Years.Front Immunol. 2022 May 26;13:900509. doi: 10.3389/fimmu.2022.900509. eCollection 2022. Front Immunol. 2022. PMID: 35720310 Free PMC article. Review.
-
The Correlation between Subolesin-Reactive Epitopes and Vaccine Efficacy.Vaccines (Basel). 2022 Aug 16;10(8):1327. doi: 10.3390/vaccines10081327. Vaccines (Basel). 2022. PMID: 36016215 Free PMC article.
-
Proteomic Analyses of Acinetobacter baumannii Clinical Isolates to Identify Drug Resistant Mechanism.Front Cell Infect Microbiol. 2021 Feb 24;11:625430. doi: 10.3389/fcimb.2021.625430. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33718272 Free PMC article.
-
Designing Multi-Antigen Vaccines Against Acinetobacter baumannii Using Systemic Approaches.Front Immunol. 2021 Apr 16;12:666742. doi: 10.3389/fimmu.2021.666742. eCollection 2021. Front Immunol. 2021. PMID: 33936107 Free PMC article.
-
Genetic Engineering of Klebsiella pneumoniae ATCC 25955 for Bioconjugate Vaccine Applications.Microorganisms. 2023 May 17;11(5):1321. doi: 10.3390/microorganisms11051321. Microorganisms. 2023. PMID: 37317295 Free PMC article.
References
-
- Alvarez-Hamelin J. I., Dall'Asta L., Barrat A., Vespignani A. (2005). K-core decomposition of internet graphs: hierarchies, self-similarity and measurement biases. arXiv. 10.3934/nhm.2008.3.371 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

