Identification of Salivary Microbiota and Its Association With Host Inflammatory Mediators in Periodontitis
- PMID: 31281801
- PMCID: PMC6598052
- DOI: 10.3389/fcimb.2019.00216
Identification of Salivary Microbiota and Its Association With Host Inflammatory Mediators in Periodontitis
Abstract
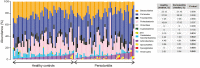
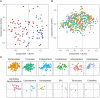
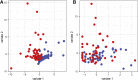
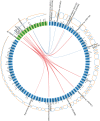
Periodontitis is a microbial-induced chronic inflammatory disease, which may not only result in tooth loss, but can also contribute to the development of various systemic diseases. The transition from healthy to diseased periodontium depends on microbial dysbiosis and impaired host immune response. Although periodontitis is a common disease as well as associated with various systemic inflammatory conditions, the taxonomic profiling of the salivary microbiota in periodontitis and its association with host immune and inflammatory mediators has not been reported. Therefore, the aim of this study was to identify key pathogens and their potential interaction with the host's inflammatory mediators in saliva samples for periodontitis risk assessment. The microbial 16S rRNA gene sequencing and the levels of inflammatory mediators were performed in saliva samples from patients with chronic periodontitis and periodontally healthy control subjects. The salivary microbial community composition differed significantly between patients with chronic periodontitis and healthy controls. Our analyses identified a number of microbes, including bacteria assigned to Eubacterium saphenum, Tannerella forsythia, Filifactor alocis, Streptococcus mitis/parasanguinis, Parvimonas micra, Prevotella sp., Phocaeicola sp., and Fretibacterium sp. as more abundant in periodontitis, compared to healthy controls. In samples from healthy individuals, we identified Campylobacter concisus, and Veillonella sp. as more abundant. Integrative analysis of the microbiota and inflammatory mediators/cytokines revealed associations that included positive correlations between the pathogens Treponema sp. and Selenomas sp. and the cytokines chitinase 3-like 1, sIL-6Rα, sTNF-R1, and gp130/sIL-6Rβ. In addition, a negative correlation was identified between IL-10 and Filifactor alocis. Our results reveal distinct and disease-specific patterns of salivary microbial composition between patients with periodontitis and healthy controls, as well as significant correlations between microbiota and host-mediated inflammatory cytokines. The positive correlations between the pathogens Treponema sp. and Selenomas sp. and the cytokines chitinase 3-like 1, sIL-6Rα, sTNF-R1, and gp130/sIL-6Rβ might have the future potential to serve as a combined bacteria-host salivary biomarker panel for diagnosis of the chronic infectious disease periodontitis. However, further studies are required to determine the capacity of these microbes and inflammatory mediators as a salivary biomarker panel for periodontitis.
Keywords: 16S rRNA sequencing; cytokines; inflammatory mediators; microbiome; microbiota; periodontitis; saliva.
Figures



References
-
- Belstrom D., Constancias F., Liu Y., Yang L., Drautz-Moses D. I., Schuster S. C., et al. (2017a). Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes 3:23. 10.1038/s41522-017-0031-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

