Analysis of Potential Genes and Pathways Involved in the Pathogenesis of Acne by Bioinformatics
- PMID: 31281837
- PMCID: PMC6590534
- DOI: 10.1155/2019/3739086
Analysis of Potential Genes and Pathways Involved in the Pathogenesis of Acne by Bioinformatics
Abstract
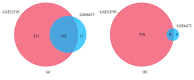
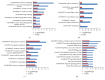
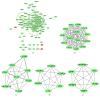
Acne is the eighth most frequent disease worldwide. Inflammatory response runs through all stages of acne. It is complicated and is involved in innate and adaptive immunity. This study aimed to explore the candidate genes and their relative signaling pathways in inflammatory acne using data mining analysis. Microarray data GSE6475 and GSE53795, including 18 acne lesion tissues and 18 matched normal skin tissues, were obtained. Differentially expressed genes (DEGs) were filtered and subjected to functional and pathway enrichment analyses. Protein-protein interaction (PPI) network and module analyses were also performed based on the DEGs. In this work, 154 common DEGs, including 145 upregulated and 9 downregulated, were obtained from two microarray profiles. Gene Ontology and pathway enrichment of DEGs were clustered using significant enrichment analysis. A PPI network containing 110 nodes/DEGs was constructed, and 31 hub genes were obtained. Four modules in the PPI network, which mainly participated in chemokine signaling pathway, cytokine-cytokine receptor interaction, and Fc gamma R-mediated phagocytosis, were extracted. In conclusion, aberrant DEGs and pathways involved in acne pathogenesis were identified using bioinformatic analysis. The DEGs included FPR2, ITGB2, CXCL8, C3AR1, CXCL1, FCER1G, LILRB2, PTPRC, SAA1, CCR2, ICAM1, and FPR1, and the pathways included chemokine signaling pathway, cytokine-cytokine receptor interaction, and Fc gamma R-mediated phagocytosis. This study could serve as a basis for further understanding the pathogenesis and potential therapeutic targets of inflammatory acne.
Figures
Similar articles
-
Identification of Genes and Pathways Associated with Acne Using Integrated Bioinformatics Methods.Dermatology. 2019;235(6):445-455. doi: 10.1159/000502203. Epub 2019 Aug 27. Dermatology. 2019. PMID: 31454825
-
Integrative bioinformatics and experimental validation of hub genetic markers in acne vulgaris: Toward personalized diagnostic and therapeutic strategies.J Cosmet Dermatol. 2024 May;23(5):1777-1799. doi: 10.1111/jocd.16152. Epub 2024 Jan 24. J Cosmet Dermatol. 2024. PMID: 38268224
-
Identification and verification immune-related regulatory network in acne.Int Immunopharmacol. 2020 Dec;89(Pt B):107083. doi: 10.1016/j.intimp.2020.107083. Epub 2020 Oct 14. Int Immunopharmacol. 2020. PMID: 33068860
-
Identification of key genes and pathways in pelvic organ prolapse based on gene expression profiling by bioinformatics analysis.Arch Gynecol Obstet. 2018 May;297(5):1323-1332. doi: 10.1007/s00404-018-4745-1. Epub 2018 Mar 15. Arch Gynecol Obstet. 2018. PMID: 29546564
-
Is there a relationship between psoriasis and hepatitis C? A meta-analysis and bioinformatics investigation.Virol J. 2021 Jul 2;18(1):135. doi: 10.1186/s12985-021-01606-z. Virol J. 2021. PMID: 34215260 Free PMC article.
Cited by
-
Gene variants associated with acne vulgaris presentation and severity: a systematic review and meta-analysis.BMC Med Genomics. 2021 Apr 13;14(1):103. doi: 10.1186/s12920-021-00953-8. BMC Med Genomics. 2021. PMID: 33849530 Free PMC article.
-
Integrated Analysis of Key Genes and Pathways Involved in Nonalcoholic Steatohepatitis Improvement After Roux-en-Y Gastric Bypass Surgery.Front Endocrinol (Lausanne). 2021 Feb 2;11:611213. doi: 10.3389/fendo.2020.611213. eCollection 2020. Front Endocrinol (Lausanne). 2021. PMID: 33603714 Free PMC article.
-
Review-Current Concepts in Inflammatory Skin Diseases Evolved by Transcriptome Analysis: In-Depth Analysis of Atopic Dermatitis and Psoriasis.Int J Mol Sci. 2020 Jan 21;21(3):699. doi: 10.3390/ijms21030699. Int J Mol Sci. 2020. PMID: 31973112 Free PMC article. Review.
-
Altered Gene Expression in Acne Vulgaris Patients Treated by Oral Isotretinoin: A Preliminary Study.Pharmgenomics Pers Med. 2020 Sep 15;13:385-395. doi: 10.2147/PGPM.S250969. eCollection 2020. Pharmgenomics Pers Med. 2020. PMID: 32982373 Free PMC article.
-
Putative Genes and Pathways Involved in the Acne Treatment of Isotretinoin via Microarray Data Analyses.Biomed Res Int. 2020 Jun 29;2020:5842795. doi: 10.1155/2020/5842795. eCollection 2020. Biomed Res Int. 2020. PMID: 32685503 Free PMC article.
References
-
- Contassot E., Beer H.-D., French L. E. Interleukin-1, inflammasomes, autoinflammation and the skin. Swiss Medical Weekly. 2012;142, article w13590 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

