Human Systems Biology and Metabolic Modelling: A Review-From Disease Metabolism to Precision Medicine
- PMID: 31281846
- PMCID: PMC6590590
- DOI: 10.1155/2019/8304260
Human Systems Biology and Metabolic Modelling: A Review-From Disease Metabolism to Precision Medicine
Abstract
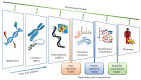
In cell and molecular biology, metabolism is the only system that can be fully simulated at genome scale. Metabolic systems biology offers powerful abstraction tools to simulate all known metabolic reactions in a cell, therefore providing a snapshot that is close to its observable phenotype. In this review, we cover the 15 years of human metabolic modelling. We show that, although the past five years have not experienced large improvements in the size of the gene and metabolite sets in human metabolic models, their accuracy is rapidly increasing. We also describe how condition-, tissue-, and patient-specific metabolic models shed light on cell-specific changes occurring in the metabolic network, therefore predicting biomarkers of disease metabolism. We finally discuss current challenges and future promising directions for this research field, including machine/deep learning and precision medicine. In the omics era, profiling patients and biological processes from a multiomic point of view is becoming more common and less expensive. Starting from multiomic data collected from patients and N-of-1 trials where individual patients constitute different case studies, methods for model-building and data integration are being used to generate patient-specific models. Coupled with state-of-the-art machine learning methods, this will allow characterizing each patient's disease phenotype and delivering precision medicine solutions, therefore leading to preventative medicine, reduced treatment, and in silico clinical trials.
Figures

References
-
- Yurkovich J. T., Palsson B. O. Solving puzzles with missing pieces: The power of systems biology [Point of View] Proceedings of the IEEE. 2016;104(1):2–7. doi: 10.1109/JPROC.2015.2505338. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

