Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily
- PMID: 31283204
- PMCID: PMC6677139
- DOI: 10.1021/acs.biochem.9b00341
Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily
Abstract
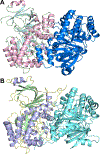
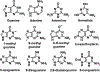
Guanine deaminase is a metabolic enzyme, found in all forms of life, which catalyzes the conversion of guanine to xanthine. Despite the availability of several crystal structures, the molecular determinants of substrate orientation and mechanism remain to be elucidated for the amidohydrolase family of guanine deaminase enzymes. Here, we report the crystal structures of Escherichia coli and Saccharomyces cerevisiae guanine deaminase enzymes (EcGuaD and Gud1, respectively), both members of the amidohydrolase superfamily. EcGuaD and Gud1 retain the overall TIM barrel tertiary structure conserved among amidohydrolase enzymes. Both proteins also possess a single zinc cation with trigonal bipyrimidal coordination geometry within their active sites. We also determined a liganded structure of Gud1 bound to the product, xanthine. Analysis of this structure, along with kinetic data of native and site-directed mutants of EcGuaD, identifies several key residues that are responsible for substrate recognition and catalysis. In addition, after a small library of compounds had been screened, two guanine derivatives, 8-azaguanine and 1-methylguanine, were identified as EcGuaD substrates. Interestingly, both EcGuaD and Gud1 also exhibit secondary ammeline deaminase activity. Overall, this work details key structural features of substrate recognition and catalysis of the amidohydrolase family of guanine deaminase enzymes in support of our long-term goal to engineer these enzymes with altered activity and substrate specificity.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
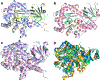
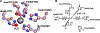


Similar articles
-
Bacterial ammeline metabolism via guanine deaminase.J Bacteriol. 2010 Feb;192(4):1106-12. doi: 10.1128/JB.01243-09. Epub 2009 Dec 18. J Bacteriol. 2010. PMID: 20023034 Free PMC article.
-
Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase.Biochemistry. 2013 Nov 12;52(45):8106-14. doi: 10.1021/bi400818e. Epub 2013 Oct 28. Biochemistry. 2013. PMID: 24083949
-
Rescue of the orphan enzyme isoguanine deaminase.Biochemistry. 2011 Jun 28;50(25):5555-7. doi: 10.1021/bi200680y. Epub 2011 Jun 7. Biochemistry. 2011. PMID: 21604715 Free PMC article.
-
Aspartase/fumarase superfamily: a common catalytic strategy involving general base-catalyzed formation of a highly stabilized aci-carboxylate intermediate.Biochemistry. 2012 May 29;51(21):4237-43. doi: 10.1021/bi300430j. Epub 2012 May 15. Biochemistry. 2012. PMID: 22551392 Review.
-
Structural and catalytic diversity within the amidohydrolase superfamily.Biochemistry. 2005 May 3;44(17):6383-91. doi: 10.1021/bi047326v. Biochemistry. 2005. PMID: 15850372 Review.
Cited by
-
Valacyclovir and Acyclovir Are Substrates of the Guanine Deaminase Cytosolic PSD-95 Interactor (Cypin).Proteins. 2025 Feb;93(2):430-440. doi: 10.1002/prot.26740. Epub 2024 Aug 29. Proteins. 2025. PMID: 39210666 Free PMC article.
-
Binding asymmetry and conformational studies of the AtGSDA dimer.Comput Struct Biotechnol J. 2023 Nov 4;21:5515-5522. doi: 10.1016/j.csbj.2023.11.004. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38022696 Free PMC article.
-
Structure of Klebsiella pneumoniae adenosine monophosphate nucleosidase.PLoS One. 2022 Oct 20;17(10):e0275023. doi: 10.1371/journal.pone.0275023. eCollection 2022. PLoS One. 2022. PMID: 36264993 Free PMC article.
-
A drought-responsive rice amidohydrolase is the elusive plant guanine deaminase with the potential to modulate the epigenome.Physiol Plant. 2021 Aug;172(4):1853-1866. doi: 10.1111/ppl.13392. Epub 2021 Apr 1. Physiol Plant. 2021. PMID: 33749847 Free PMC article.
-
Retracing the Rapid Evolution of an Herbicide-Degrading Enzyme by Protein Engineering.ACS Catal. 2023 Nov 17;13(23):15558-15571. doi: 10.1021/acscatal.3c04010. ACS Catal. 2023. PMID: 38567019 Free PMC article.
References
-
- Raushel FM (2016) Finding homes for orphan enzymes, Perspectives in Science 9, 3–7.
-
- Holm L, and Sander C (1997) An evolutionary treasure: unification of a broad set of amidohydrolases related to urease, Proteins 28, 72–82. - PubMed
-
- Seibert CM, and Raushel FM (2005) Structural and catalytic diversity within the amidohydrolase superfamily, Biochemistry 44, 6383–6391. - PubMed
-
- Liaw SH, Chang YJ, Lai CT, Chang HC, and Chang GG (2004) Crystal structure of Bacillus subtilis guanine deaminase: the first domain-swapped structure in the cytidine deaminase superfamily, The Journal of biological chemistry 279, 35479–35485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

