Alu RNA Modulates the Expression of Cell Cycle Genes in Human Fibroblasts
- PMID: 31284509
- PMCID: PMC6651528
- DOI: 10.3390/ijms20133315
Alu RNA Modulates the Expression of Cell Cycle Genes in Human Fibroblasts
Abstract
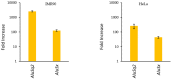
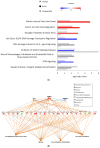
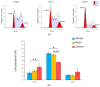
Alu retroelements, whose retrotransposition requires prior transcription by RNA polymerase III to generate Alu RNAs, represent the most numerous non-coding RNA (ncRNA) gene family in the human genome. Alu transcription is generally kept to extremely low levels by tight epigenetic silencing, but it has been reported to increase under different types of cell perturbation, such as viral infection and cancer. Alu RNAs, being able to act as gene expression modulators, may be directly involved in the mechanisms determining cellular behavior in such perturbed states. To directly address the regulatory potential of Alu RNAs, we generated IMR90 fibroblasts and HeLa cell lines stably overexpressing two slightly different Alu RNAs, and analyzed genome-wide the expression changes of protein-coding genes through RNA-sequencing. Among the genes that were upregulated or downregulated in response to Alu overexpression in IMR90, but not in HeLa cells, we found a highly significant enrichment of pathways involved in cell cycle progression and mitotic entry. Accordingly, Alu overexpression was found to promote transition from G1 to S phase, as revealed by flow cytometry. Therefore, increased Alu RNA may contribute to sustained cell proliferation, which is an important factor of cancer development and progression.
Keywords: Alu retrotransposons; cell cycle; non-coding RNA.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Berger A., Strub K. Multiple Roles of Alu-Related Noncoding RNAs. Prog. Mol. Subcell. Biol. 2011;51:119–146. - PubMed
-
- Versteeg R., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

