The Biological Axis of Protein Arginine Methylation and Asymmetric Dimethylarginine
- PMID: 31284549
- PMCID: PMC6651691
- DOI: 10.3390/ijms20133322
The Biological Axis of Protein Arginine Methylation and Asymmetric Dimethylarginine
Abstract
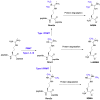
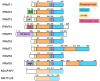
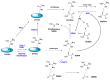
Protein post-translational modifications (PTMs) in eukaryotic cells play important roles in the regulation of functionalities of the proteome and in the tempo-spatial control of cellular processes. Most PTMs enact their regulatory functions by affecting the biochemical properties of substrate proteins such as altering structural conformation, protein-protein interaction, and protein-nucleic acid interaction. Amid various PTMs, arginine methylation is widespread in all eukaryotic organisms, from yeasts to humans. Arginine methylation in many situations can drastically or subtly affect the interactions of substrate proteins with their partnering proteins or nucleic acids, thus impacting major cellular programs. Recently, arginine methylation has become an important regulator of the formation of membrane-less organelles inside cells, a phenomenon of liquid-liquid phase separation (LLPS), through altering π-cation interactions. Another unique feature of arginine methylation lies in its impact on cellular physiology through its downstream amino acid product, asymmetric dimethylarginine (ADMA). Accumulation of ADMA in cells and in the circulating bloodstream is connected with endothelial dysfunction and a variety of syndromes of cardiovascular diseases. Herein, we review the current knowledge and understanding of protein arginine methylation in regards to its canonical function in direct protein regulation, as well as the biological axis of protein arginine methylation and ADMA biology.
Keywords: ADMA; PRMT; arginine methylation; metabolism; nitric oxide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Asymmetric dimethylarginine (ADMA), symmetric dimethylarginine (SDMA) and homoarginine (hArg): the ADMA, SDMA and hArg paradoxes.Cardiovasc Diabetol. 2018 Jan 4;17(1):1. doi: 10.1186/s12933-017-0656-x. Cardiovasc Diabetol. 2018. PMID: 29301528 Free PMC article.
-
Increased protein arginine methylation in chronic hypoxia: role of protein arginine methyltransferases.Am J Respir Cell Mol Biol. 2006 Oct;35(4):436-43. doi: 10.1165/rcmb.2006-0097OC. Epub 2006 May 11. Am J Respir Cell Mol Biol. 2006. PMID: 16690984
-
ADMA metabolism and clearance.Vasc Med. 2005 Jul;10 Suppl 1:S73-81. doi: 10.1191/1358863x05vm597oa. Vasc Med. 2005. PMID: 16444872 Review.
-
From arginine methylation to ADMA: a novel mechanism with therapeutic potential in chronic lung diseases.BMC Pulm Med. 2009 Jan 29;9:5. doi: 10.1186/1471-2466-9-5. BMC Pulm Med. 2009. PMID: 19178698 Free PMC article. Review.
-
Asymmetric dimethylarginine, an endogenous NOS inhibitor, is actively metabolized in rat erythrocytes.Biosci Biotechnol Biochem. 2012;76(7):1334-42. doi: 10.1271/bbb.120086. Epub 2012 Jul 7. Biosci Biotechnol Biochem. 2012. PMID: 22785485
Cited by
-
Dysregulation of the Nitric Oxide/Dimethylarginine Pathway in Hypoxic Pulmonary Vasoconstriction-Molecular Mechanisms and Clinical Significance.Front Med (Lausanne). 2022 Feb 17;9:835481. doi: 10.3389/fmed.2022.835481. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35252268 Free PMC article. Review.
-
The Importance of a New Cardiovascular Risk Factor - Asymmetric Dimethylarginine.Maedica (Bucur). 2020 Sep;15(3):373-375. doi: 10.26574/maedica.2020.15.3.373. Maedica (Bucur). 2020. PMID: 33312254 Free PMC article.
-
CARM1 Regulates AMPK Signaling in Skeletal Muscle.iScience. 2020 Nov 2;23(11):101755. doi: 10.1016/j.isci.2020.101755. eCollection 2020 Nov 20. iScience. 2020. PMID: 33241200 Free PMC article.
-
Deep structural insights into RNA-binding disordered protein regions.Wiley Interdiscip Rev RNA. 2022 Sep;13(5):e1714. doi: 10.1002/wrna.1714. Epub 2022 Jan 30. Wiley Interdiscip Rev RNA. 2022. PMID: 35098694 Free PMC article. Review.
-
Altered L-Arginine Metabolic Pathways in Gastric Cancer: Potential Therapeutic Targets and Biomarkers.Biomolecules. 2021 Jul 23;11(8):1086. doi: 10.3390/biom11081086. Biomolecules. 2021. PMID: 34439753 Free PMC article.
References
-
- Bulau P., Zakrzewicz D., Kitowska K., Wardega B., Kreuder J., Eickelberg O. Quantitative assessment of arginine methylation in free versus protein-incorporated amino acids in vitro and in vivo using protein hydrolysis and high-performance liquid chromatography. BioTechniques. 2006;40:305–310. doi: 10.2144/000112081. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

