The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation
- PMID: 31284572
- PMCID: PMC6651423
- DOI: 10.3390/ijms20133328
The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation
Abstract
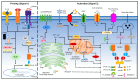
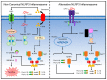
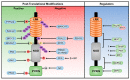
The NLRP3 inflammasome is a critical component of the innate immune system that mediates caspase-1 activation and the secretion of proinflammatory cytokines IL-1β/IL-18 in response to microbial infection and cellular damage. However, the aberrant activation of the NLRP3 inflammasome has been linked with several inflammatory disorders, which include cryopyrin-associated periodic syndromes, Alzheimer's disease, diabetes, and atherosclerosis. The NLRP3 inflammasome is activated by diverse stimuli, and multiple molecular and cellular events, including ionic flux, mitochondrial dysfunction, and the production of reactive oxygen species, and lysosomal damage have been shown to trigger its activation. How NLRP3 responds to those signaling events and initiates the assembly of the NLRP3 inflammasome is not fully understood. In this review, we summarize our current understanding of the mechanisms of NLRP3 inflammasome activation by multiple signaling events, and its regulation by post-translational modifications and interacting partners of NLRP3.
Keywords: Ionic flux; Lysosomal damage; Mitochondrial dysfunction; NLRP3 inflammasome; NLRP3 regulators; Post-translational modification; Priming; ROS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

