Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973
- PMID: 31284644
- PMCID: PMC6651687
- DOI: 10.3390/ijms20133336
Identification and Characterization of Preferred DNA-Binding Sites for the Thermus thermophilus HB8 Transcriptional Regulator TTHA0973
Abstract
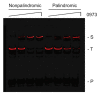
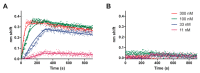
Advances in genomic sequencing have allowed the identification of a multitude of genes encoding putative transcriptional regulatory proteins. Lacking, often, is a fuller understanding of the biological roles played by these proteins, the genes they regulate or regulon. Conventionally this is achieved through a genetic approach involving putative transcription factor gene manipulation and observations of changes in an organism's transcriptome. However, such an approach is not always feasible or can yield misleading findings. Here, we describe a biochemistry-centric approach, involving identification of preferred DNA-binding sequences for the Thermus thermophilus HB8 transcriptional repressor TTHA0973 using the selection method Restriction Endonuclease Protection, Selection and Amplification (REPSA), massively parallel sequencing, and bioinformatic analyses. We identified a consensus TTHA0973 recognition sequence of 5'-AACnAACGTTnGTT-3' that exhibited nanomolar binding affinity. This sequence was mapped to several sites within the T. thermophilus HB8 genome, a subset of which corresponded to promoter regions regulating genes involved in phenylacetic acid degradation. These studies further demonstrate the utility of a biochemistry-centric approach for the facile identification of potential biological functions for orphan transcription factors in a variety of organisms.
Keywords: bioinformatics; biolayer interferometry (BLI); electrophoretic mobility shift assay (EMSA); extremophile; type IIS restriction endonuclease.
Conflict of interest statement
The authors declare no conflict of interest. In addition, the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

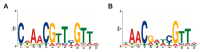

References
-
- Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A., et al. RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016;44:D133–D143. doi: 10.1093/nar/gkv1156. - DOI - PMC - PubMed
-
- Oshima T., Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. Nov., a non-sporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 1974;24:102–112. doi: 10.1099/00207713-24-1-102. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

