Altered Nuclear Export Signal Recognition as a Driver of Oncogenesis
- PMID: 31285298
- PMCID: PMC6774834
- DOI: 10.1158/2159-8290.CD-19-0298
Altered Nuclear Export Signal Recognition as a Driver of Oncogenesis
Abstract
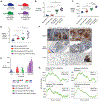
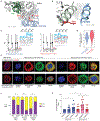
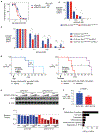
Altered expression of XPO1, the main nuclear export receptor in eukaryotic cells, has been observed in cancer, and XPO1 has been a focus of anticancer drug development. However, mechanistic evidence for cancer-specific alterations in XPO1 function is lacking. Here, genomic analysis of 42,793 cancers identified recurrent and previously unrecognized mutational hotspots in XPO1. XPO1 mutations exhibited striking lineage specificity, with enrichment in a variety of B-cell malignancies, and introduction of single amino acid substitutions in XPO1 initiated clonal, B-cell malignancy in vivo. Proteomic characterization identified that mutant XPO1 altered the nucleocytoplasmic distribution of hundreds of proteins in a sequence-specific manner that promoted oncogenesis. XPO1 mutations preferentially sensitized cells to inhibitors of nuclear export, providing a biomarker of response to this family of drugs. These data reveal a new class of oncogenic alteration based on change-of-function mutations in nuclear export signal recognition and identify therapeutic targets based on altered nucleocytoplasmic trafficking. SIGNIFICANCE: Here, we identify that heterozygous mutations in the main nuclear exporter in eukaryotic cells, XPO1, are positively selected in cancer and promote the initiation of clonal B-cell malignancies. XPO1 mutations alter nuclear export signal recognition in a sequence-specific manner and sensitize cells to compounds in clinical development inhibiting XPO1 function.This article is highlighted in the In This Issue feature, p. 1325.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures




Comment in
-
Delivering to the wrong address.Nat Rev Cancer. 2019 Oct;19(10):542-543. doi: 10.1038/s41568-019-0200-2. Nat Rev Cancer. 2019. PMID: 31439948 No abstract available.
References
-
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–50. - PubMed
-
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–11. - PubMed
-
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

