Adaptation by naïve CD4+ T cells to self-antigen-dependent TCR signaling induces functional heterogeneity and tolerance
- PMID: 31285342
- PMCID: PMC6660790
- DOI: 10.1073/pnas.1904096116
Adaptation by naïve CD4+ T cells to self-antigen-dependent TCR signaling induces functional heterogeneity and tolerance
Abstract
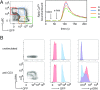
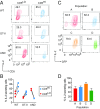
Naïve CD4+ T cells experience weak T cell receptor (TCR) signals induced by self-peptides presented by MHC II. To investigate how these "basal" TCR signals influence responses to agonist TCR ligand stimulation, we analyzed naïve CD4+ cells expressing varying amounts of CD5, Ly6C, and Nur77-GFP, markers that reflect the strength of basal TCR signaling. Phenotypic analyses indicate that the broadest range of basal TCR signal strength can be visualized by a combination of Nur77-GFP and Ly6C. A range of basal TCR signaling is detectable even in populations that express identical TCRs. Whereas moderate basal TCR signal strength correlates with higher IL-2 secretion at early time points following TCR stimulation, weak basal TCR signaling correlated with higher IL-2 secretion at later time points. We identify a population of Nur77-GFPHI Ly6C- cells that could not be reliably marked by either of CD5, Ly6C, or Nur77-GFP alone. These cells experience the strongest basal TCR signaling, consistently produce less IL-2, and express PD-1 and markers associated with anergy, such as Grail and Cbl-b. We propose that adaptation to the strength of basal TCR signaling drives the phenotypic and functional heterogeneity of naïve CD4+ cells.
Keywords: CD5; Nur77; T cell activation; T cell anergy; basal TCR signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Strong Basal/Tonic TCR Signals Are Associated with Negative Regulation of Naive CD4+ T Cells.Immunohorizons. 2022 Sep 13;6(9):671-683. doi: 10.4049/immunohorizons.2200051. Immunohorizons. 2022. PMID: 36100367
-
IL-2 Modulates the TCR Signaling Threshold for CD8 but Not CD4 T Cell Proliferation on a Single-Cell Level.J Immunol. 2017 Mar 15;198(6):2445-2456. doi: 10.4049/jimmunol.1601453. Epub 2017 Feb 3. J Immunol. 2017. PMID: 28159902 Free PMC article.
-
Functional heterogeneity and adaptation of naive T cells in response to tonic TCR signals.Curr Opin Immunol. 2021 Dec;73:43-49. doi: 10.1016/j.coi.2021.09.007. Epub 2021 Oct 12. Curr Opin Immunol. 2021. PMID: 34653787 Free PMC article. Review.
-
Reporters of TCR signaling identify arthritogenic T cells in murine and human autoimmune arthritis.Proc Natl Acad Sci U S A. 2019 Sep 10;116(37):18517-18527. doi: 10.1073/pnas.1904271116. Epub 2019 Aug 27. Proc Natl Acad Sci U S A. 2019. PMID: 31455730 Free PMC article.
-
Narcissistic T cells: reactivity to self makes a difference.FEBS J. 2021 Mar;288(6):1778-1788. doi: 10.1111/febs.15498. Epub 2020 Aug 10. FEBS J. 2021. PMID: 32738029 Review.
Cited by
-
Adapting T Cell Receptor Ligand Discrimination Capability via LAT.Front Immunol. 2021 Apr 16;12:673196. doi: 10.3389/fimmu.2021.673196. eCollection 2021. Front Immunol. 2021. PMID: 33936119 Free PMC article. Review.
-
The perception and response of T cells to a changing environment are based on the law of initial value.Sci Signal. 2022 May 31;15(736):eabj9842. doi: 10.1126/scisignal.abj9842. Epub 2022 May 31. Sci Signal. 2022. PMID: 35639856 Free PMC article. Review.
-
Cross-Reactivity with Self-Antigen Tunes the Functional Potential of Naive B Cells Specific for Foreign Antigens.J Immunol. 2020 Feb 1;204(3):498-509. doi: 10.4049/jimmunol.1900799. Epub 2019 Dec 27. J Immunol. 2020. PMID: 31882518 Free PMC article.
-
TCR signaling promotes formation of an STS1-Cbl-b complex with pH-sensitive phosphatase activity that suppresses T cell function in acidic environments.Immunity. 2023 Dec 12;56(12):2682-2698.e9. doi: 10.1016/j.immuni.2023.11.010. Immunity. 2023. PMID: 38091950 Free PMC article.
-
Cathepsin L-dependent positive selection shapes clonal composition and functional fitness of CD4+ T cells.Nat Immunol. 2025 Jul;26(7):1127-1138. doi: 10.1038/s41590-025-02182-y. Epub 2025 Jun 13. Nat Immunol. 2025. PMID: 40514418 Free PMC article.
References
-
- Schluns K. S., Kieper W. C., Jameson S. C., Lefrançois L., Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 1, 426–432 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

