A reliable Epstein-Barr Virus classification based on phylogenomic and population analyses
- PMID: 31285478
- PMCID: PMC6614506
- DOI: 10.1038/s41598-019-45986-3
A reliable Epstein-Barr Virus classification based on phylogenomic and population analyses
Abstract
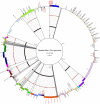
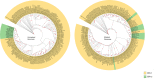
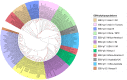
The Epstein-Barr virus (EBV) infects more than 90% of the human population, playing a key role in the origin and progression of malignant and non-malignant diseases. Many attempts have been made to classify EBV according to clinical or epidemiological information; however, these classifications show frequent incongruences. For instance, they use a small subset of genes for sorting strains but fail to consider the enormous genomic variability and abundant recombinant regions present in the EBV genome. These could lead to diversity overestimation, alter the tree topology and misinterpret viral types when classified, therefore, a reliable EBV phylogenetic classification is needed to minimize recombination signals. Recombination events occur 2.5-times more often than mutation events, suggesting that recombination has a much stronger impact than mutation in EBV genomic diversity, detected within common ancestral node positions. The Hierarchical Bayesian Analysis of Population Structure (hierBAPS) resulted in the differentiation of 12 EBV populations showed seven monophyletic and five paraphyletic. The populations identified were related to geographic location, of which three populations (EBV-p1/Asia/GC, EBV-p2/Asia II/Tumors and EBV-p4/China/NPC) were related to tumor development. Therefore, we proposed a new consistent and non-simplistic EBV classification, beneficial in minimizing the recombination signal in the phylogeny reconstruction, investigating geography relationship and even infer associations to human diseases. These EBV classifications could also be useful in developing diagnostic applications or defining which strains need epidemiological surveillance.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Early Epstein-Barr Virus Genomic Diversity and Convergence toward the B95.8 Genome in Primary Infection.J Virol. 2018 Jan 2;92(2):e01466-17. doi: 10.1128/JVI.01466-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29093087 Free PMC article.
-
Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection.J Virol. 2015 May;89(10):5222-37. doi: 10.1128/JVI.03614-14. Epub 2015 Mar 18. J Virol. 2015. PMID: 25787276 Free PMC article.
-
Comprehensive Evolutionary Analysis of Complete Epstein-Barr Virus Genomes from Argentina and Other Geographies.Viruses. 2021 Jun 18;13(6):1172. doi: 10.3390/v13061172. Viruses. 2021. PMID: 34207433 Free PMC article.
-
Phylogenetic comparison of Epstein-Barr virus genomes.J Microbiol. 2018 Aug;56(8):525-533. doi: 10.1007/s12275-018-8039-x. Epub 2018 Jun 14. J Microbiol. 2018. PMID: 29948828 Review.
-
Epstein-Barr virus strain variation and cancer.Cancer Sci. 2019 Apr;110(4):1132-1139. doi: 10.1111/cas.13954. Epub 2019 Feb 21. Cancer Sci. 2019. PMID: 30697862 Free PMC article. Review.
Cited by
-
Genotype characterization of Epstein-Barr virus among adults living with human immunodeficiency virus in Ethiopia.Front Microbiol. 2023 Oct 31;14:1270824. doi: 10.3389/fmicb.2023.1270824. eCollection 2023. Front Microbiol. 2023. PMID: 38029140 Free PMC article.
-
Exploring the Genetic Diversity of Epstein-Barr Virus among Patients with Gastric Cancer in Southern Chile.Int J Mol Sci. 2023 Jul 10;24(14):11276. doi: 10.3390/ijms241411276. Int J Mol Sci. 2023. PMID: 37511034 Free PMC article.
-
Genetic Patterns Found in the Nuclear Localization Signals (NLSs) Associated with EBV-1 and EBV-2 Provide New Insights into Their Contribution to Different Cell-Type Specificities.Cancers (Basel). 2021 May 24;13(11):2569. doi: 10.3390/cancers13112569. Cancers (Basel). 2021. PMID: 34073836 Free PMC article.
-
The 'Oma's of the Gammas-Cancerogenesis by γ-Herpesviruses.Viruses. 2024 Dec 17;16(12):1928. doi: 10.3390/v16121928. Viruses. 2024. PMID: 39772235 Free PMC article. Review.
-
EBNA1 Upregulates P53-Inhibiting Genes in Burkitt's Lymphoma Cell Line.Rep Biochem Mol Biol. 2023 Jan;11(4):672-683. doi: 10.52547/rbmb.11.4.672. Rep Biochem Mol Biol. 2023. PMID: 37131894 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

