Genome assembly of Nannochloropsis oceanica provides evidence of host nucleus overthrow by the symbiont nucleus during speciation
- PMID: 31286066
- PMCID: PMC6610115
- DOI: 10.1038/s42003-019-0500-9
Genome assembly of Nannochloropsis oceanica provides evidence of host nucleus overthrow by the symbiont nucleus during speciation
Abstract
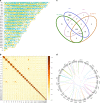
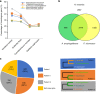
The species of the genus Nannochloropsis are unique in their maintenance of a nucleus-plastid continuum throughout their cell cycle, non-motility and asexual reproduction. These characteristics should have been endorsed in their gene assemblages (genomes). Here we show that N. oceanica has a genome of 29.3 Mb consisting of 32 pseudochromosomes and containing 7,330 protein-coding genes; and the host nucleus may have been overthrown by an ancient red alga symbiont nucleus during speciation through secondary endosymbiosis. In addition, N. oceanica has lost its flagella and abilities to undergo meiosis and sexual reproduction, and adopted a genome reduction strategy during speciation. We propose that N. oceanica emerged through the active fusion of a host protist and a photosynthesizing ancient red alga and the symbiont nucleus became dominant over the host nucleus while the chloroplast was wrapped by two layers of endoplasmic reticulum. Our findings evidenced an alternative speciation pathway of eukaryotes.
Keywords: Evolution; Genomics.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures

Similar articles
-
Genomic footprints of a cryptic plastid endosymbiosis in diatoms.Science. 2009 Jun 26;324(5935):1724-6. doi: 10.1126/science.1172983. Science. 2009. PMID: 19556510
-
Chromera velia, endosymbioses and the rhodoplex hypothesis--plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages).Genome Biol Evol. 2014 Mar;6(3):666-84. doi: 10.1093/gbe/evu043. Genome Biol Evol. 2014. PMID: 24572015 Free PMC article.
-
Algal genes in the closest relatives of animals.Mol Biol Evol. 2010 Dec;27(12):2879-89. doi: 10.1093/molbev/msq175. Epub 2010 Jul 13. Mol Biol Evol. 2010. PMID: 20627874
-
What was the real contribution of endosymbionts to the eukaryotic nucleus? Insights from photosynthetic eukaryotes.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a016014. doi: 10.1101/cshperspect.a016014. Cold Spring Harb Perspect Biol. 2014. PMID: 24984774 Free PMC article. Review.
-
Integration of plastids with their hosts: Lessons learned from dinoflagellates.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10247-54. doi: 10.1073/pnas.1421380112. Epub 2015 May 20. Proc Natl Acad Sci U S A. 2015. PMID: 25995366 Free PMC article. Review.
Cited by
-
Metabolomics Reveals the Impact of Overexpression of Cytosolic Fructose-1,6-Bisphosphatase on Photosynthesis and Growth in Nannochloropsis gaditana.Int J Mol Sci. 2024 Jun 20;25(12):6800. doi: 10.3390/ijms25126800. Int J Mol Sci. 2024. PMID: 38928505 Free PMC article.
-
Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte.Viruses. 2021 Sep 15;13(9):1837. doi: 10.3390/v13091837. Viruses. 2021. PMID: 34578418 Free PMC article.
-
Engineering Nannochloropsis oceanica for the production of diterpenoid compounds.mLife. 2023 Dec 26;2(4):428-437. doi: 10.1002/mlf2.12097. eCollection 2023 Dec. mLife. 2023. PMID: 38818264 Free PMC article.
-
Phenotypic Analysis and Molecular Characterization of Enlarged Cell Size Mutant in Nannochloropsis oceanica.Int J Mol Sci. 2023 Sep 2;24(17):13595. doi: 10.3390/ijms241713595. Int J Mol Sci. 2023. PMID: 37686401 Free PMC article.
-
Toward the Exploitation of Sustainable Green Factory: Biotechnology Use of Nannochloropsis spp.Biology (Basel). 2024 Apr 25;13(5):292. doi: 10.3390/biology13050292. Biology (Basel). 2024. PMID: 38785776 Free PMC article. Review.
References
-
- Sukenik A, Carmeli Y, Berner T. Regulation of fatty acid composition by irradiance level in the Eustigmatophyte Nannochloropsis sp. J. Phycol. 1989;25:686–692. doi: 10.1111/j.0022-3646.1989.00686.x. - DOI
-
- Boussiba S, et al. Lipid and biomass production by the halotolerant microalga Nannochloropsis salina. Biomass. 1987;12:37–47. doi: 10.1016/0144-4565(87)90006-0. - DOI
-
- Galloway RE. Selective condition and isolation of mutants in salt-tolerant, lipid-producing microalgae. J. Phycol. 1990;26:752–260. doi: 10.1111/j.0022-3646.1990.00752.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

