Identification and differential expression analysis of anthocyanin biosynthetic genes in leaf color variants of ornamental kale
- PMID: 31286853
- PMCID: PMC6615239
- DOI: 10.1186/s12864-019-5910-z
Identification and differential expression analysis of anthocyanin biosynthetic genes in leaf color variants of ornamental kale
Abstract
Background: Anthocyanins perform diverse biological functions in plants and are beneficial to human health. Leaf color is the most important trait of ornamental kale and the characteristics of changes in leaf color make it an ideal material to elucidate genetic mechanisms of anthocyanins accumulation in Brassica oleracea. To elucidate the anthocyanin distribution, metabolic profiles and differentially expressed anthocyanin biosynthetic genes between different colored accessions can pave the way for understanding the genetic regulatory mechanisms of anthocyanin biosynthesis and accumulation in ornamental kale.
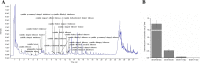
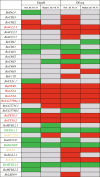
Results: In this study, anthocyanin distributions in red- and white-leaved ornamental kale accessions were determined. Thirty-four anthocyanins were detected in the red-leaved accession. The complete set of anthocyanin biosynthetic genes in the B. oleracea reference genome was identified and differential expression analysis based on RNA-seq was conducted. Eighty-one anthocyanin biosynthetic genes were identified in the B. oleracea reference genome. The expression patterns and differential expressions of these genes in different leaf types indicated that late biosynthetic genes (BoDFR1, BoANS1 and 2, and BoUGT79B1.1), positive regulatory genes (BoTTG1, BoTT8, and Bol012528), a negative regulatory gene (BoMYBL2.1), and transport genes (BoTT19.1 and BoTT19.2) may play roles in anthocyanin accumulation in ornamental kale. A genetic regulatory network of anthocyanin accumulation in ornamental kale was constructed.
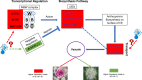
Conclusions: The distribution of pigments and anthocyanin profiles explained the leaf color phenotypes of ornamental kales. The identification of key genes and construction of genetic regulatory network in anthocyanin accumulation in ornamental kale elucidated the genetic basis of leaf color variants. These findings enhance the understanding of the genetic mechanisms and regulatory network of anthocyanin accumulation in B. oleracea, and provide a theoretical basis for breeding new cultivars of Brassica vegetables with enhanced ornamental and nutritional value.
Keywords: Anthocyanin accumulation; Anthocyanin biosynthetic genes; Brassica oleracea; Color variants; Differentially expressed genes; Ornamental kale; Regulatory network.
Conflict of interest statement
All of the authors declare that there are no personal, professional or financial relationships that could potentially be construed as a conflict of interest.
Figures



References
-
- Anders S, Huber W. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL); 2012.
-
- Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol. 2005;8(3):272–279. - PubMed
-
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol. 2008;26(11):1301–1308. - PubMed
MeSH terms
Substances
Grants and funding
- 31501752/Young Scientists Fund
- KJCX20170413/The Science and Technology Innovation Capacity Building Projects of the Beijing Academy of Agriculture and Forestry Sciences
- P172001038/State Administration of Foreign Experts Affairs
- 2015GA600004/The National Spark Program Projects of China
- GJHZ2017-2/The International Cooperation Fund of the Beijing Academy of Agriculture and Forestry Sciences
LinkOut - more resources
Full Text Sources

