Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages
- PMID: 31286919
- PMCID: PMC6615263
- DOI: 10.1186/s12870-019-1916-6
Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages
Abstract
Background: SPL (SQUAMOSA promoter binding protein-like) is a class of plant-specific transcription factors that play important roles in many growth and developmental processes, including shoot and inflorescence branching, embryonic development, signal transduction, leaf initiation, phase transition, and flower and fruit development. The SPL gene family has been identified and characterized in many species but has not been well studied in tartary buckwheat, which is an important edible and medicinal crop.
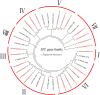
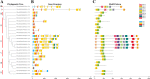
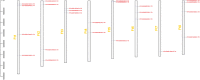
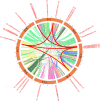
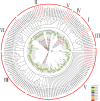
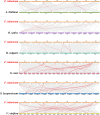
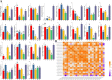
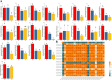
Results: In this study, 24 Fagopyrum tataricum SPL (FtSPL) genes were identified and renamed according to the chromosomal distribution of the FtSPL genes. According to the amino acid sequence of the SBP domain and gene structure, the SPL genes were divided into eight groups (group I to group VII) by phylogenetic tree analysis. A total of 10 motifs were detected in the tartary buckwheat SPL genes. The expression patterns of 23 SPL genes in different tissues and fruits at different developmental stages (green fruit stage, discoloration stage and initial maturity stage) were determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Conclusions: The tartary buckwheat genome contained 24 SPL genes, and most of the genes were expressed in different tissues. qRT-PCR showed that FtSPLs played important roles in the growth and development of tartary buckwheat, and genes that might regulate flower and fruit development were preliminarily identified. This work provides a comprehensive understanding of the SBP-box gene family in tartary buckwheat and lays a significant foundation for further studies on the functional characteristics of FtSPL genes and improvement of tartary buckwheat crops.
Keywords: Expression patterns; Fruit development; FtSPL; Genome; Tartary buckwheat.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Takanori O, Kyoko Y, Ohmi O. Two new Fagopyrum (Polygonaceae) species, F. gracilipedoides and F. jinshaense from Yunnan, China. Jpn J Genet. 2002;77(6):399–408. - PubMed
-
- Eggum BO, Kreft I, Javornik B. Chemical composition and protein quality of buckwheat ( Fagopyrum esculentum Moench ) Plant Foods Hum Nutr. 1980;30(3–4):175–179.
-
- Bonafaccia G, Marocchini M, Kreft I. Composition and technological properties of the flour and bran from common and tartary buckwheat. Food Chem. 2003;80(1):9–15.
-
- Lee CC, Shen SR, Lai YJ, Wu SC. Rutin and quercetin, bioactive compounds from tartary buckwheat, prevent liver inflammatory injury. Food Funct. 2013;4(5):794–802. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

