Characterization of swine-origin H1N1 canine influenza viruses
- PMID: 31287780
- PMCID: PMC7011970
- DOI: 10.1080/22221751.2019.1637284
Characterization of swine-origin H1N1 canine influenza viruses
Abstract
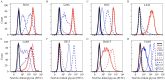
Host switch events of influenza A viruses (IAVs) continuously pose a zoonotic threat to humans. In 2013, swine-origin H1N1 IAVs emerged in dogs soon after they were detected in swine in the Guangxi province of China. This host switch was followed by multiple reassortment events between these H1N1 and previously circulating H3N2 canine IAVs (IAVs-C) in dogs. To evaluate the phenotype of these newly identified viruses, we characterized three swine-origin H1N1 IAVs-C and one reassortant H1N1 IAV-C. We found that H1N1 IAVs-C predominantly bound to human-type receptors, efficiently transmitted via direct contact in guinea pigs and replicated in human lung cells. Moreover, the swine-origin H1N1 IAVs-C were lethal in mice and were transmissible by respiratory droplets in guinea pigs. Importantly, sporadic human infections with these viruses have been detected, and preexisting immunity in humans might not be sufficient to prevent infections with these new viruses. Our results show the potential of H1N1 IAVs-C to infect and transmit in humans, suggesting that these viruses should be closely monitored in the future.
Keywords: H1N1; Host switch; canine; influenza A viruses; reassortant.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

Similar articles
-
Emergence and Evolution of Novel Reassortant Influenza A Viruses in Canines in Southern China.mBio. 2018 Jun 5;9(3):e00909-18. doi: 10.1128/mBio.00909-18. mBio. 2018. PMID: 29871917 Free PMC article.
-
Zoonotic Risk, Pathogenesis, and Transmission of Avian-Origin H3N2 Canine Influenza Virus.J Virol. 2017 Oct 13;91(21):e00637-17. doi: 10.1128/JVI.00637-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28814512 Free PMC article.
-
Molecular characterization of a novel reassortant H1N2 influenza virus containing genes from the 2009 pandemic human H1N1 virus in swine from eastern China.Virus Genes. 2016 Jun;52(3):405-10. doi: 10.1007/s11262-016-1303-4. Epub 2016 Mar 15. Virus Genes. 2016. PMID: 26980674
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Isolation and genetic characterization of avian-like H1N1 and novel ressortant H1N2 influenza viruses from pigs in China.Biochem Biophys Res Commun. 2009 Aug 21;386(2):278-83. doi: 10.1016/j.bbrc.2009.05.056. Epub 2009 May 19. Biochem Biophys Res Commun. 2009. PMID: 19460353 Review.
Cited by
-
Live attenuated influenza A virus vaccines with modified NS1 proteins for veterinary use.Front Cell Infect Microbiol. 2022 Jul 22;12:954811. doi: 10.3389/fcimb.2022.954811. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35937688 Free PMC article. Review.
-
Influenza A viruses circulating in dogs: A review of the scientific literature.Open Vet J. 2022 Sep-Oct;12(5):676-687. doi: 10.5455/OVJ.2022.v12.i5.12. Epub 2022 Sep 15. Open Vet J. 2022. PMID: 36589407 Free PMC article. Review.
-
Decitabine Increases the Transcription of RIG-I Gene to Suppress the Replication of Feline Calicivirus and Canine Influenza Virus.Microorganisms. 2025 Jan 13;13(1):143. doi: 10.3390/microorganisms13010143. Microorganisms. 2025. PMID: 39858911 Free PMC article.
-
Genetic characterization and pathogenicity of a Eurasian avian-like H1N1 swine influenza reassortant virus.Virol J. 2022 Dec 2;19(1):205. doi: 10.1186/s12985-022-01936-6. Virol J. 2022. PMID: 36461007 Free PMC article.
-
A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial.Nat Med. 2021 Jan;27(1):106-114. doi: 10.1038/s41591-020-1118-7. Epub 2020 Dec 7. Nat Med. 2021. PMID: 33288923 Clinical Trial.
References
-
- Li C, Chen H.. Enhancement of influenza virus transmission by gene reassortment. Curr Top MicrobiolImmunol. 2014;385:185–204. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
