Novel antibiotics effective against gram-positive and -negative multi-resistant bacteria with limited resistance
- PMID: 31287812
- PMCID: PMC6615598
- DOI: 10.1371/journal.pbio.3000337
Novel antibiotics effective against gram-positive and -negative multi-resistant bacteria with limited resistance
Abstract
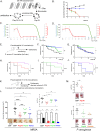
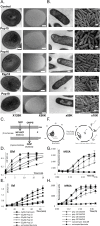
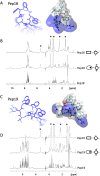
Antibiotics are a medical wonder, but an increasing frequency of resistance among most human pathogens is rendering them ineffective. If this trend continues, the consequences for public health and for the general community could be catastrophic. The current clinical pipeline, however, is very limited and is dominated by derivatives of established classes, the "me too" compounds. Here, we have exploited our recent identification of a bacterial toxin to transform it into antibiotics active on multidrug-resistant (MDR) gram-positive and -negative bacterial pathogens. We generated a new family of peptidomimetics-cyclic heptapseudopeptides-inspired from a natural bacterial peptide. Out of the 4 peptides studied, 2 are effective against methicillin-resistant Staphylococcus aureus (MRSA) in mild and severe sepsis mouse models without exhibiting toxicity on human erythrocytes and kidney cells, zebrafish embryos, and mice. These new compounds are safe at their active doses and above, without nephrotoxicity. Efficacy was also demonstrated against Pseudomonas aeruginosa and MRSA in a mouse skin infection model. Importantly, these compounds did not result in resistance after serial passages for 2 weeks and 4 or 6 days' exposure in mice. Activity of heptapseudopeptides was explained by the ability of unnatural amino acids to strengthen dynamic association with bacterial lipid bilayers and to induce membrane permeability, leading to bacterial death. Based on structure determination, we showed that cationic domains surrounded by an extended hydrophobic core could improve bactericidal activity. Because 2 peptide analogs, Pep 16 and Pep19, are effective against both MRSA and P. aeruginosa in severe sepsis and skin infection models, respectively, we believe that these peptidomimetics are promising lead candidates for drug development. We have identified potential therapeutic agents that can provide alternative treatments against antimicrobial resistance. Because the compounds are potential leads for therapeutic development, the next step is to start phase I clinical trials.
Conflict of interest statement
Cyclic peptides Pep15, Pep16, Pep18, and Pep19 are covered under European patent application #EP14191238.6, “Cyclic antimicrobial pseudopeptides and uses thereof.”
Figures
Similar articles
-
Cationic antimicrobial peptides: alternatives and/or adjuvants to antibiotics active against methicillin-resistant Staphylococcus aureus and multidrug-resistant Pseudomonas aeruginosa.BMC Microbiol. 2019 Mar 8;19(1):54. doi: 10.1186/s12866-019-1416-8. BMC Microbiol. 2019. PMID: 30849936 Free PMC article.
-
The Neutrally Charged Diarylurea Compound PQ401 Kills Antibiotic-Resistant and Antibiotic-Tolerant Staphylococcus aureus.mBio. 2020 Jun 30;11(3):e01140-20. doi: 10.1128/mBio.01140-20. mBio. 2020. PMID: 32605985 Free PMC article.
-
Antibiotics Enhance Prevention and Eradication Efficacy of Cathodic-Voltage-Controlled Electrical Stimulation against Titanium-Associated Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa Biofilms.mSphere. 2019 May 1;4(3):e00178-19. doi: 10.1128/mSphere.00178-19. mSphere. 2019. PMID: 31043516 Free PMC article.
-
Delafloxacin: a novel fluoroquinolone with activity against methicillin-resistant Staphylococcus aureus (MRSA) and Pseudomonas aeruginosa.Expert Rev Anti Infect Ther. 2018 Jul;16(7):523-530. doi: 10.1080/14787210.2018.1489721. Epub 2018 Jun 26. Expert Rev Anti Infect Ther. 2018. PMID: 29911455 Review.
-
Antibiotic considerations in the treatment of multidrug-resistant (MDR) pathogens: a case-based review.J Hosp Med. 2009 Jul;4(6):E8-15. doi: 10.1002/jhm.505. J Hosp Med. 2009. PMID: 19670375 Review.
Cited by
-
Novel temporin L antimicrobial peptides: promoting self-assembling by lipidic tags to tackle superbugs.J Enzyme Inhib Med Chem. 2020 Dec;35(1):1751-1764. doi: 10.1080/14756366.2020.1819258. J Enzyme Inhib Med Chem. 2020. PMID: 32957844 Free PMC article.
-
Synergistic Activity of Pep16, a Promising New Antibacterial Pseudopeptide against Multidrug-Resistant Organisms, in Combination with Colistin against Multidrug-Resistant Escherichia coli, In Vitro and in a Murine Peritonitis Model.Antibiotics (Basel). 2023 Jan 3;12(1):81. doi: 10.3390/antibiotics12010081. Antibiotics (Basel). 2023. PMID: 36671282 Free PMC article.
-
Bacterial Type I Toxins: Folding and Membrane Interactions.Toxins (Basel). 2021 Jul 14;13(7):490. doi: 10.3390/toxins13070490. Toxins (Basel). 2021. PMID: 34357962 Free PMC article. Review.
-
Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity.Mar Drugs. 2020 Dec 10;18(12):630. doi: 10.3390/md18120630. Mar Drugs. 2020. PMID: 33321943 Free PMC article.
-
Mechanism of action of sprG1-encoded type I toxins in Staphylococcus aureus: from membrane alterations to mesosome-like structures formation and bacterial lysis.Front Microbiol. 2023 Oct 3;14:1275849. doi: 10.3389/fmicb.2023.1275849. eCollection 2023. Front Microbiol. 2023. PMID: 37854335 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

