Super-resolution microscopy reveals how histone tail acetylation affects DNA compaction within nucleosomes in vivo
- PMID: 31287868
- PMCID: PMC6895258
- DOI: 10.1093/nar/gkz593
Super-resolution microscopy reveals how histone tail acetylation affects DNA compaction within nucleosomes in vivo
Abstract
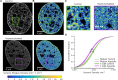
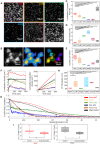
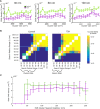
Chromatin organization is crucial for regulating gene expression. Previously, we showed that nucleosomes form groups, termed clutches. Clutch size correlated with the pluripotency grade of mouse embryonic stem cells and human induced pluripotent stem cells. Recently, it was also shown that regions of the chromatin containing activating epigenetic marks were composed of small and dispersed chromatin nanodomains with lower DNA density compared to the larger silenced domains. Overall, these results suggest that clutch size may regulate DNA packing density and gene activity. To directly test this model, we carried out 3D, two-color super-resolution microscopy of histones and DNA with and without increased histone tail acetylation. Our results showed that lower percentage of DNA was associated with nucleosome clutches in hyperacetylated cells. We further showed that the radius and compaction level of clutch-associated DNA decreased in hyperacetylated cells, especially in regions containing several neighboring clutches. Importantly, this change was independent of clutch size but dependent on the acetylation state of the clutch. Our results directly link the epigenetic state of nucleosome clutches to their DNA packing density. Our results further provide in vivo support to previous in vitro models that showed a disruption of nucleosome-DNA interactions upon hyperacetylation.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Cremer T., Cremer M., Cremer C.. The 4D nucleome: genome compartmentalization in an evolutionary context. Biochemistry (Biokhimiia). 2018; 83:313–325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

