Reconstitution of Microtubule Nucleation In Vitro Reveals Novel Roles for Mzt1
- PMID: 31287970
- PMCID: PMC6616311
- DOI: 10.1016/j.cub.2019.05.058
Reconstitution of Microtubule Nucleation In Vitro Reveals Novel Roles for Mzt1
Erratum in
-
Reconstitution of Microtubule Nucleation In Vitro Reveals Novel Roles for Mzt1.Curr Biol. 2023 May 8;33(9):1864. doi: 10.1016/j.cub.2023.04.030. Curr Biol. 2023. PMID: 37160082 Free PMC article. No abstract available.
Abstract
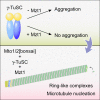
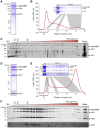
Microtubule (MT) nucleation depends on the γ-tubulin complex (γ-TuC), in which multiple copies of the heterotetrameric γ-tubulin small complex (γ-TuSC) associate to form a ring-like structure (in metazoans, γ-tubulin ring complex; γ-TuRC) [1-7]. Additional conserved regulators of the γ-TuC include the small protein Mzt1 (MOZART1 in human; GIP1/1B and GIP2/1A in plants) [8-13] and proteins containing a Centrosomin Motif 1 (CM1) domain [10, 14-19]. Many insights into γ-TuC regulators have come from in vivo analysis in fission yeast Schizosaccharomyces pombe. The S. pombe CM1 protein Mto1 recruits the γ-TuC to microtubule-organizing centers (MTOCs) [14, 20-22], and analysis of Mto1[bonsai], a truncated version of Mto1 that cannot localize to MTOCs, has shown that Mto1 also has a role in γ-TuC activation [23]. S. pombe Mzt1 interacts with γ-TuSC and is essential for γ-TuC function and localization to MTOCs [11, 12]. However, the mechanisms by which Mzt1 functions remain unclear. Here we describe reconstitution of MT nucleation using purified recombinant Mto1[bonsai], the Mto1 partner protein Mto2, γ-TuSC, and Mzt1. Multiple copies of the six proteins involved coassemble to form a 34-40S ring-like "MGM" holocomplex that is a potent MT nucleator in vitro. Using purified MGM and subcomplexes, we investigate the role of Mzt1 in MT nucleation. Our results suggest that Mzt1 is critical to stabilize Alp6, the S. pombe homolog of human γ-TuSC protein GCP3, in an "interaction-competent" form within the γ-TuSC. This is essential for MGM to become a functional nucleator.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



Similar articles
-
Activation of the γ-tubulin complex by the Mto1/2 complex.Curr Biol. 2014 Apr 14;24(8):896-903. doi: 10.1016/j.cub.2014.03.006. Epub 2014 Apr 3. Curr Biol. 2014. PMID: 24704079 Free PMC article.
-
Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo.J Cell Sci. 2008 Dec 1;121(Pt 23):3971-80. doi: 10.1242/jcs.038414. Epub 2008 Nov 11. J Cell Sci. 2008. PMID: 19001497 Free PMC article.
-
Fission yeast MOZART1/Mzt1 is an essential γ-tubulin complex component required for complex recruitment to the microtubule organizing center, but not its assembly.Mol Biol Cell. 2013 Sep;24(18):2894-906. doi: 10.1091/mbc.E13-05-0235. Epub 2013 Jul 24. Mol Biol Cell. 2013. PMID: 23885124 Free PMC article.
-
Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence.Trends Cell Biol. 2015 May;25(5):296-307. doi: 10.1016/j.tcb.2014.12.002. Epub 2014 Dec 24. Trends Cell Biol. 2015. PMID: 25544667 Review.
-
Acentrosomal microtubule nucleation in higher plants.Int Rev Cytol. 2002;220:257-89. doi: 10.1016/s0074-7696(02)20008-x. Int Rev Cytol. 2002. PMID: 12224551 Review.
Cited by
-
Distribution of γ-tubulin ring complex at the Schizosaccharomyces pombe spindle pole.MicroPubl Biol. 2021 Sep 16;2021:10.17912/micropub.biology.000464. doi: 10.17912/micropub.biology.000464. eCollection 2021. MicroPubl Biol. 2021. PMID: 35622906 Free PMC article.
-
The cryo-EM structure of a γ-TuSC elucidates architecture and regulation of minimal microtubule nucleation systems.Nat Commun. 2020 Nov 11;11(1):5705. doi: 10.1038/s41467-020-19456-8. Nat Commun. 2020. PMID: 33177498 Free PMC article.
-
Klp2-mediated Rsp1-Mto1 colocalization inhibits microtubule-dependent microtubule assembly in fission yeast.Sci Adv. 2025 Jan 3;11(1):eadq0670. doi: 10.1126/sciadv.adq0670. Epub 2025 Jan 3. Sci Adv. 2025. PMID: 39752482 Free PMC article.
-
Structure of the native γ-tubulin ring complex capping spindle microtubules.Nat Struct Mol Biol. 2024 Jul;31(7):1134-1144. doi: 10.1038/s41594-024-01281-y. Epub 2024 Apr 12. Nat Struct Mol Biol. 2024. PMID: 38609662 Free PMC article.
-
γ-Tubulin Complexes and Fibrillar Arrays: Two Conserved High Molecular Forms with Many Cellular Functions.Cells. 2021 Apr 1;10(4):776. doi: 10.3390/cells10040776. Cells. 2021. PMID: 33915825 Free PMC article. Review.
References
-
- Kollman J.M., Merdes A., Mourey L., Agard D.A. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011;12:709–721. - PMC - PubMed
- Kollman, J.M., Merdes, A., Mourey, L., and Agard, D.A. (2011). Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709-721. - PMC - PubMed
-
- Lin T.C., Neuner A., Schiebel E. Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends Cell Biol. 2015;25:296–307. - PubMed
- Lin, T.C., Neuner, A., and Schiebel, E. (2015). Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends Cell Biol. 25, 296-307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

