Clustered protocadherins methylation alterations in cancer
- PMID: 31288858
- PMCID: PMC6617643
- DOI: 10.1186/s13148-019-0695-0
Clustered protocadherins methylation alterations in cancer
Abstract
Background: Clustered protocadherins (PCDHs) map in tandem at human chromosome 5q31 and comprise three multi-genes clusters: α-, β- and γ-PCDH. The expression of this cluster consists of a complex mechanism involving DNA hub formation through DNA-CCTC binding factor (CTCF) interaction. Methylation alterations can affect this interaction, leading to transcriptional dysregulation. In cancer, clustered PCDHs undergo a mechanism of long-range epigenetic silencing by hypermethylation.
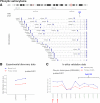
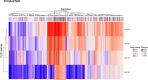
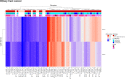
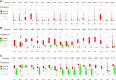
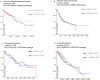
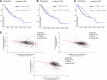
Results: In this study, we detected frequent methylation alterations at CpG islands associated to these clustered PCDHs in all the solid tumours analysed (colorectal, gastric and biliary tract cancers, pilocytic astrocytoma), but not hematologic neoplasms such as chronic lymphocytic leukemia. Importantly, several altered CpG islands were associated with CTCF binding sites. Interestingly, our analysis revealed a hypomethylation event in pilocytic astrocytoma, suggesting that in neuronal tissue, where PCDHs are highly expressed, these genes become hypomethylated in this type of cancer. On the other hand, in tissues where PCDHs are lowly expressed, these CpG islands are targeted by DNA methylation. In fact, PCDH-associated CpG islands resulted hypermethylated in gastrointestinal tumours.
Conclusions: Our study highlighted a strong alteration of the clustered PCDHs methylation pattern in the analysed solid cancers and suggested these methylation aberrations in the CpG islands associated with PCDH genes as powerful diagnostic biomarkers.
Keywords: BTC; Biliary tract cancer; CLL; CRA; CRC; CTCF; Cancer methylation alteration; Chronic lymphocytic leukemia; Clustered PCDH; Colorectal adenoma; Colorectal carcinoma; CpG islands; GC; Gastric cancer; LGG; Low grade glioma; PA; Pilocytic astrocytoma.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14893–14898. doi: 10.1073/pnas.1004526107. - DOI - PMC - PubMed
-
- Dallosso AR, Oster B, Greenhough A, Thorsen K, Curry TJ, Owen C, et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. 2012;31(40):4409–4419. doi: 10.1038/onc.2011.609. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

