Ubiquinone Biosynthesis over the Entire O2 Range: Characterization of a Conserved O2-Independent Pathway
- PMID: 31289180
- PMCID: PMC6747719
- DOI: 10.1128/mBio.01319-19
Ubiquinone Biosynthesis over the Entire O2 Range: Characterization of a Conserved O2-Independent Pathway
Abstract
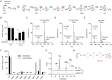
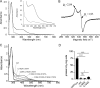
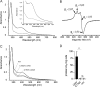
Most bacteria can generate ATP by respiratory metabolism, in which electrons are shuttled from reduced substrates to terminal electron acceptors, via quinone molecules like ubiquinone. Dioxygen (O2) is the terminal electron acceptor of aerobic respiration and serves as a co-substrate in the biosynthesis of ubiquinone. Here, we characterize a novel, O2-independent pathway for the biosynthesis of ubiquinone. This pathway relies on three proteins, UbiT (YhbT), UbiU (YhbU), and UbiV (YhbV). UbiT contains an SCP2 lipid-binding domain and is likely an accessory factor of the biosynthetic pathway, while UbiU and UbiV (UbiU-UbiV) are involved in hydroxylation reactions and represent a novel class of O2-independent hydroxylases. We demonstrate that UbiU-UbiV form a heterodimer, wherein each protein binds a 4Fe-4S cluster via conserved cysteines that are essential for activity. The UbiT, -U, and -V proteins are found in alpha-, beta-, and gammaproteobacterial clades, including several human pathogens, supporting the widespread distribution of a previously unrecognized capacity to synthesize ubiquinone in the absence of O2 Together, the O2-dependent and O2-independent ubiquinone biosynthesis pathways contribute to optimizing bacterial metabolism over the entire O2 range.IMPORTANCE In order to colonize environments with large O2 gradients or fluctuating O2 levels, bacteria have developed metabolic responses that remain incompletely understood. Such adaptations have been recently linked to antibiotic resistance, virulence, and the capacity to develop in complex ecosystems like the microbiota. Here, we identify a novel pathway for the biosynthesis of ubiquinone, a molecule with a key role in cellular bioenergetics. We link three uncharacterized genes of Escherichia coli to this pathway and show that the pathway functions independently from O2 In contrast, the long-described pathway for ubiquinone biosynthesis requires O2 as a substrate. In fact, we find that many proteobacteria are equipped with the O2-dependent and O2-independent pathways, supporting that they are able to synthesize ubiquinone over the entire O2 range. Overall, we propose that the novel O2-independent pathway is part of the metabolic plasticity developed by proteobacteria to face various environmental O2 levels.
Keywords: bioenergetics; facultative anaerobes; hydroxylases; iron-sulfur; oxygen; peptidase U32; proteobacteria; quinones; respiration; ubiquinone.
Copyright © 2019 Pelosi et al.
Figures




Similar articles
-
The O2-independent pathway of ubiquinone biosynthesis is essential for denitrification in Pseudomonas aeruginosa.J Biol Chem. 2020 Jul 3;295(27):9021-9032. doi: 10.1074/jbc.RA120.013748. Epub 2020 May 14. J Biol Chem. 2020. PMID: 32409583 Free PMC article.
-
Role of the Escherichia coli ubiquinone-synthesizing UbiUVT pathway in adaptation to changing respiratory conditions.mBio. 2023 Aug 31;14(4):e0329822. doi: 10.1128/mbio.03298-22. Epub 2023 Jun 7. mBio. 2023. PMID: 37283518 Free PMC article.
-
The UbiK protein is an accessory factor necessary for bacterial ubiquinone (UQ) biosynthesis and forms a complex with the UQ biogenesis factor UbiJ.J Biol Chem. 2017 Jul 14;292(28):11937-11950. doi: 10.1074/jbc.M117.789164. Epub 2017 May 30. J Biol Chem. 2017. PMID: 28559279 Free PMC article.
-
Biosynthesis and physiology of coenzyme Q in bacteria.Biochim Biophys Acta. 2014 Jul;1837(7):1004-11. doi: 10.1016/j.bbabio.2014.01.015. Epub 2014 Jan 28. Biochim Biophys Acta. 2014. PMID: 24480387 Review.
-
Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors.Biochim Biophys Acta. 1997 Jul 4;1320(3):217-34. doi: 10.1016/s0005-2728(97)00034-0. Biochim Biophys Acta. 1997. PMID: 9230919 Review.
Cited by
-
Regulation of Reactive Oxygen Species Promotes Growth and Carotenoid Production Under Autotrophic Conditions in Rhodobacter sphaeroides.Front Microbiol. 2022 Feb 28;13:847757. doi: 10.3389/fmicb.2022.847757. eCollection 2022. Front Microbiol. 2022. PMID: 35295297 Free PMC article.
-
Diversification of Ubiquinone Biosynthesis via Gene Duplications, Transfers, Losses, and Parallel Evolution.Mol Biol Evol. 2023 Oct 4;40(10):msad219. doi: 10.1093/molbev/msad219. Mol Biol Evol. 2023. PMID: 37788637 Free PMC article.
-
Effects of colonization-associated gene yqiC on global transcriptome, cellular respiration, and oxidative stress in Salmonella Typhimurium.J Biomed Sci. 2022 Dec 1;29(1):102. doi: 10.1186/s12929-022-00885-0. J Biomed Sci. 2022. PMID: 36457101 Free PMC article.
-
The O2-independent pathway of ubiquinone biosynthesis is essential for denitrification in Pseudomonas aeruginosa.J Biol Chem. 2020 Jul 3;295(27):9021-9032. doi: 10.1074/jbc.RA120.013748. Epub 2020 May 14. J Biol Chem. 2020. PMID: 32409583 Free PMC article.
-
The PhoPQ Two-Component System Is the Major Regulator of Cell Surface Properties, Stress Responses and Plant-Derived Substrate Utilisation During Development of Pectobacterium versatile-Host Plant Pathosystems.Front Microbiol. 2021 Jan 15;11:621391. doi: 10.3389/fmicb.2020.621391. eCollection 2020. Front Microbiol. 2021. PMID: 33519782 Free PMC article.
References
-
- Wallace N, Zani A, Abrams E, Sun Y. 2016. The impact of oxygen on bacterial enteric pathogens, p 179–204. In Sariaslani S, Gadd GM (ed), Advances in applied microbiology, vol 95 Elsevier Academic Press Inc, San Diego, CA. - PubMed
-
- Bettenbrock K, Bai H, Ederer M, Green J, Hellingwerf KJ, Holcombe M, Kunz S, Rolfe MD, Sanguinetti G, Sawodny O, Sharma P, Steinsiek S, Poole RK. 2014. Towards a systems level understanding of the oxygen response of Escherichia coli. Adv Microb Syst Biol 64:65–114. doi:10.1016/B978-0-12-800143-1.00002-6. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

