Genomic selection on breeding time in a wild bird population
- PMID: 31289689
- PMCID: PMC6591552
- DOI: 10.1002/evl3.103
Genomic selection on breeding time in a wild bird population
Abstract
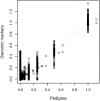
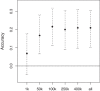
Artificial selection experiments are a powerful tool in evolutionary biology. Selecting individuals based on multimarker genotypes (genomic selection) has several advantages over phenotype-based selection but has, so far, seen very limited use outside animal and plant breeding. Genomic selection depends on the markers tagging the causal loci that underlie the selected trait. Because the number of necessary markers depends, among other factors, on effective population size, genomic selection may be in practice not feasible in wild populations as most wild populations have much higher effective population sizes than domesticated populations. However, the current possibilities of cost-effective high-throughput genotyping could overcome this limitation and thereby make it possible to apply genomic selection also in wild populations. Using a unique dataset of about 2000 wild great tits (Parus major), a small passerine bird, genotyped on a 650 k SNP chip we calculated genomic breeding values for egg-laying date using the so-called GBLUP approach. In this approach, the pedigree-based relatedness matrix of an "animal model," a special form of the mixed model, is replaced by a marker-based relatedness matrix. Using the marker-based relatedness matrix, the model seemed better able to disentangle genetic and permanent environmental effects. We calculated the accuracy of genomic breeding values by correlating them to the phenotypes of individuals whose phenotypes were excluded from the analysis when estimating the genomic breeding values. The obtained accuracy was about 0.20, with very little effect of the used genomic relatedness estimator but a strong effect of the number of SNPs. The obtained accuracy is lower than typically seen in domesticated species but considerable for a trait with low heritability (∼0.2) as avian breeding time. Our results show that genomic selection is possible also in wild populations with potentially many applications, which we discuss here.
Keywords: Animal model; GBLUP; genomic breeding values; phenology; quantitative genetic.
Figures

Similar articles
-
Accuracy of genomic selection for a sib-evaluated trait using identity-by-state and identity-by-descent relationships.Genet Sel Evol. 2015 Feb 25;47(1):9. doi: 10.1186/s12711-014-0084-2. Genet Sel Evol. 2015. PMID: 25888184 Free PMC article.
-
Increased accuracy of artificial selection by using the realized relationship matrix.Genet Res (Camb). 2009 Feb;91(1):47-60. doi: 10.1017/S0016672308009981. Genet Res (Camb). 2009. PMID: 19220931
-
Use of a Bayesian model including QTL markers increases prediction reliability when test animals are distant from the reference population.J Dairy Sci. 2019 Aug;102(8):7237-7247. doi: 10.3168/jds.2018-15815. Epub 2019 May 31. J Dairy Sci. 2019. PMID: 31155255
-
Invited review: Genomic selection in dairy cattle: progress and challenges.J Dairy Sci. 2009 Feb;92(2):433-43. doi: 10.3168/jds.2008-1646. J Dairy Sci. 2009. PMID: 19164653 Review.
-
Genomic selection.J Anim Breed Genet. 2007 Dec;124(6):323-30. doi: 10.1111/j.1439-0388.2007.00702.x. J Anim Breed Genet. 2007. PMID: 18076469 Review.
Cited by
-
Using genomic prediction to detect microevolutionary change of a quantitative trait.Proc Biol Sci. 2022 May 11;289(1974):20220330. doi: 10.1098/rspb.2022.0330. Epub 2022 May 11. Proc Biol Sci. 2022. PMID: 35538786 Free PMC article.
-
Exploration of tissue-specific gene expression patterns underlying timing of breeding in contrasting temperature environments in a song bird.BMC Genomics. 2019 Sep 2;20(1):693. doi: 10.1186/s12864-019-6043-0. BMC Genomics. 2019. PMID: 31477015 Free PMC article.
-
What processes must we understand to forecast regional-scale population dynamics?Proc Biol Sci. 2020 Dec 9;287(1940):20202219. doi: 10.1098/rspb.2020.2219. Epub 2020 Dec 9. Proc Biol Sci. 2020. PMID: 33290672 Free PMC article.
-
The estimation of additive genetic variance of body size in a wild passerine is sensitive to the method used to estimate relatedness among the individuals.Ecol Evol. 2024 Feb 13;14(2):e10981. doi: 10.1002/ece3.10981. eCollection 2024 Feb. Ecol Evol. 2024. PMID: 38352200 Free PMC article.
-
Correlational selection in the age of genomics.Nat Ecol Evol. 2021 May;5(5):562-573. doi: 10.1038/s41559-021-01413-3. Epub 2021 Apr 15. Nat Ecol Evol. 2021. PMID: 33859374 Review.
References
-
- Abzhanov, A. , Kuo W. P., Hartmann C., Grant B. R., Grant P. R., and Tabin C. J.. 2006. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature 442:563–567. - PubMed
-
- Abzhanov, A. , Protas M., Grant B. R., Grant P. R., and Tabin C. J.. 2004. Bmp4 and morphological variation of beaks in Darwin's finches. Science 305:1462–1465. - PubMed
-
- Aitken, S. N. , and Whitlock M. C.. 2013. Assisted gene flow to facilitate local adaptation to climate change. Ann. Rev. Ecol. Evol. Syst. 44:367–388.
-
- Barson, N. J. , Aykanat T., Hindar K., Baranski M., Bolstad G. H., Fiske P., et al. 2015. Sex‐dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 528:405–408. - PubMed
-
- Barton, N. H. , Etheridge A. M., and Véber A.. 2017. The infinitesimal model: definition, derivation, and implications. Theor. Pop. Biol. 118:50–73. - PubMed

