Targeting KRAS Mutant Lung Cancer Cells with siRNA-Loaded Bovine Serum Albumin Nanoparticles
- PMID: 31289919
- PMCID: PMC7611029
- DOI: 10.1007/s11095-019-2665-9
Targeting KRAS Mutant Lung Cancer Cells with siRNA-Loaded Bovine Serum Albumin Nanoparticles
Abstract
Purpose: KRAS is the most frequently mutated gene in human cancers. Despite its direct involvement in malignancy and intensive effort, direct inhibition of KRAS via pharmacological inhibitors has been challenging. RNAi induced knockdown using siRNAs against mutant KRAS alleles offers a promising tool for selective therapeutic silencing in KRAS-mutant lung cancers. However, the major bottleneck for clinical translation is the lack of efficient biocompatible siRNA carrier systems.
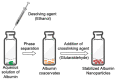
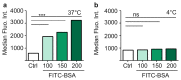
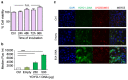
Methods: Bovine serum albumin (BSA) nanoparticles were prepared by desolvation method to deliver siRNA targeting the KRAS G12S mutation. The BSA nanoparticles were characterized with respect to their size, zeta potential, encapsulation efficiency and nucleic acid release. Nanoparticle uptake, cellular distribution of nucleic acids, cytotoxicity and gene knock down to interfere with cancer hallmarks, uncontrolled proliferation and migration, were evaluated in KRAS G12S mutant A459 cells, a lung adenocarcinoma cell line.
Results: BSA nanoparticles loaded with siRNA resulted in nanoparticles smaller than 200 nm in diameter and negative zeta potentials, displaying optimal characteristics for in vivo application. Encapsulating and protecting the siRNA payload well, the nanoparticles enabled transport to A549 cells in vitro, could evade endosomal entrapment and mediated significant sequence-specific KRAS knockdown, resulting in reduced cell growth of siRNA transfected lung cancer cells.
Conclusions: BSA nanoparticles loaded with mutant specific siRNA are a promising therapeutic approach for KRAS-mutant cancers.
Keywords: BSA; KRAS; lung cancer; siRNA delivery.
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

