Cooler: scalable storage for Hi-C data and other genomically labeled arrays
- PMID: 31290943
- PMCID: PMC8205516
- DOI: 10.1093/bioinformatics/btz540
Cooler: scalable storage for Hi-C data and other genomically labeled arrays
Abstract
Motivation: Most existing coverage-based (epi)genomic datasets are one-dimensional, but newer technologies probing interactions (physical, genetic, etc.) produce quantitative maps with two-dimensional genomic coordinate systems. Storage and computational costs mount sharply with data resolution when such maps are stored in dense form. Hence, there is a pressing need to develop data storage strategies that handle the full range of useful resolutions in multidimensional genomic datasets by taking advantage of their sparse nature, while supporting efficient compression and providing fast random access to facilitate development of scalable algorithms for data analysis.
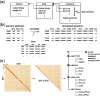
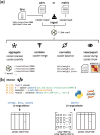
Results: We developed a file format called cooler, based on a sparse data model, that can support genomically labeled matrices at any resolution. It has the flexibility to accommodate various descriptions of the data axes (genomic coordinates, tracks and bin annotations), resolutions, data density patterns and metadata. Cooler is based on HDF5 and is supported by a Python library and command line suite to create, read, inspect and manipulate cooler data collections. The format has been adopted as a standard by the NIH 4D Nucleome Consortium.
Availability and implementation: Cooler is cross-platform, BSD-licensed and can be installed from the Python package index or the bioconda repository. The source code is maintained on Github at https://github.com/mirnylab/cooler.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Abadi D.J. et al. (2008) Column-stores vs. row-stores: how different are they really? In: Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data, SIGMOD ’08, pp. 967–980. ACM, New York, NY, USA.
-
- Collette A. (2013) Python and HDF5: Unlocking Scientific Data. O’Reilly.
-
- Davies J.O. et al. (2017) How best to identify chromosomal interactions: a comparison of approaches. Nat. Methods, 14, 125. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

