The Critical Role of Codon Composition on the Translation Efficiency Robustness of the Hepatitis A Virus Capsid
- PMID: 31290967
- PMCID: PMC6735747
- DOI: 10.1093/gbe/evz146
The Critical Role of Codon Composition on the Translation Efficiency Robustness of the Hepatitis A Virus Capsid
Abstract
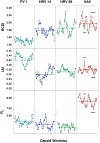
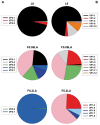
Hepatoviruses show an intriguing deviated codon usage, suggesting an evolutionary signature. Abundant and rare codons in the cellular genome are scarce in the human hepatitis A virus (HAV) genome, while intermediately abundant host codons are abundant in the virus. Genotype-phenotype maps, or fitness landscapes, are a means of representing a genotype position in sequence space and uncovering how genotype relates to phenotype and fitness. Using genotype-phenotype maps of the translation efficiency, we have shown the critical role of the HAV capsid codon composition in regulating translation and determining its robustness. Adaptation to an environmental perturbation such as the artificial induction of cellular shutoff-not naturally occurring in HAV infection-involved movements in the sequence space and dramatic changes of the translation efficiency. Capsid rare codons, including abundant and rare codons of the cellular genome, slowed down the translation efficiency in conditions of no cellular shutoff. In contrast, rare capsid codons that are abundant in the cellular genome were efficiently translated in conditions of shutoff. Capsid regions very rich in slowly translated codons adapt to shutoff through sequence space movements from positions with highly robust translation to others with diminished translation robustness. These movements paralleled decreases of the capsid physical and biological robustness, and resulted in the diversification of capsid phenotypes. The deviated codon usage of extant hepatoviruses compared with that of their hosts may suggest the occurrence of a virus ancestor with an optimized codon usage with respect to an unknown ancient host.
Keywords: codon usage; fitness landscape; genotype–phenotype maps; sequence space; translation efficiency.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Airaksinen A, Pariente N, Menéndez-Arias L, Domingo E.. 2003. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 3112:339–349. - PubMed
-
- Aldana M, Balleza E, Kauffman S, Resendiz O.. 2007. Robustness and evolvability in genetic regulatory networks. J Theor Biol. 2453:433–448. - PubMed
-
- Ancel LW, Fontana W.. 2000. Plasticity, evolvability, and modularity in RNA. J Exp Zool. 2883:242–283. - PubMed

