Transcriptome profiling of mouse brain and lung under Dip2a regulation using RNA-sequencing
- PMID: 31291246
- PMCID: PMC6619597
- DOI: 10.1371/journal.pone.0213702
Transcriptome profiling of mouse brain and lung under Dip2a regulation using RNA-sequencing
Erratum in
-
Correction: Transcriptome profiling of mouse brain and lung under Dip2a regulation using RNA-sequencing.PLoS One. 2019 Nov 14;14(11):e0225570. doi: 10.1371/journal.pone.0225570. eCollection 2019. PLoS One. 2019. PMID: 31725791 Free PMC article.
Abstract
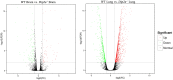
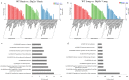
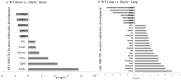
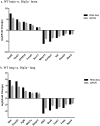
Disconnected interacting protein 2 homolog A (DIP2A) is highly expressed in nervous system and respiratory system of developing embryos. However, genes regulated by Dip2a in developing brain and lung have not been systematically studied. Transcriptome of brain and lung in embryonic 19.5 day (E19.5) were compared between wild type and Dip2a-/- mice. An average of 50 million reads per sample was mapped to the reference sequence. A total of 214 DEGs were detected in brain (82 up and 132 down) and 1900 DEGs in lung (1259 up and 641 down). GO enrichment analysis indicated that DEGs in both Brain and Lung were mainly enriched in biological processes 'DNA-templated transcription and Transcription from RNA polymerase II promoter', 'multicellular organism development', 'cell differentiation' and 'apoptotic process'. In addition, COG classification showed that both were mostly involved in 'Replication, Recombination, and Repair', 'Signal transduction and mechanism', 'Translation, Ribosomal structure and Biogenesis' and 'Transcription'. KEGG enrichment analysis showed that brain was mainly enriched in 'Thyroid cancer' pathway whereas lung in 'Complement and Coagulation Cascades' pathway. Transcription factor (TF) annotation analysis identified Zinc finger domain containing (ZF) proteins were mostly regulated in lung and brain. Interestingly, study identified genes Skor2, Gpr3711, Runx1, Erbb3, Frmd7, Fut10, Sox11, Hapln1, Tfap2c and Plxnb3 from brain that play important roles in neuronal cell maturation, differentiation, and survival; genes Hoxa5, Eya1, Errfi1, Sox11, Shh, Igf1, Ccbe1, Crh, Fgf9, Lama5, Pdgfra, Ptn, Rbp4 and Wnt7a from lung are important in lung development. Expression levels of the candidate genes were validated by qRT-PCR. Genome wide transcriptional analysis using wild type and Dip2a knockout mice in brain and lung at embryonic day 19.5 (E19.5) provided a genetic basis of molecular function of these genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Steller H, Fischbach K F, Rubin G M. Disconnected: A locus required for neuronal pathway formation in the visual system of drosophila. Cell. 1987; 50(7): 1139–1153. - PubMed
-
- Mukhopadhyay M, Pelka P, Desousa D, Kablar B, Schindler A, Rudnicki MA, et al. Cloning, genomic organization and expression pattern of a novel Drosophila gene, the disco-interacting protein 2 (dip2), and its murine homolog. Gene. 2002; 293(1): 59–65. - PubMed
-
- Tanaka M, Murakami K, Ozaki S, Imura Y, Tong XP, Watanabe T, et al. DIP2 disco-interacting protein 2 homolog A (Drosophila) is a candidate receptor for follistatin-related protein /follistatin -like1—analysis of their binding with TGF-beta super family proteins. FEBS J. 2010; 277(20): 4278–89. 10.1111/j.1742-4658.2010.07816.x - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

