Sequence and phylogenetic analysis of novel porcine parvovirus 7 isolates from pigs in Guangxi, China
- PMID: 31291362
- PMCID: PMC6619813
- DOI: 10.1371/journal.pone.0219560
Sequence and phylogenetic analysis of novel porcine parvovirus 7 isolates from pigs in Guangxi, China
Abstract
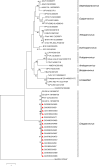
Parvoviruses are a diverse group of viruses that infect a wide range of animals and humans. In recent years, advances in molecular techniques have resulted in the identification of several novel parvoviruses in swine. In this study, porcine parvovirus 7 (PPV7) isolates from clinical samples collected in Guangxi, China, were examined to understand their molecular epidemiology and co-infection with porcine circovirus type 2 (PCV2). In this study, among the 385 pig serum samples, 105 were positive for PPV7, representing a 27.3% positive detection rate. The co-infection rate of PPV7 and PCV2 was 17.4% (67/385). Compared with the reference strains, we noted 93.9%-97.9% similarity in the NS1 gene and 87.4%-95.0% similarity in the cap gene. Interestingly, compared with the reference strains, sixteen of the PPV7 strains in this study contained an additional 3 to 15 nucleotides in the middle of the cap gene. Therefore, the Cap protein of fourteen strains encoded 474 amino acids, and the Cap protein of the other two strains encoded 470 amino acids. However, the Cap protein of the reference strain PPV7 isolate 42 encodes 469 amino acids. This is the first report of sequence variation within the cap gene, confirming an increase in the number of amino acids in the Cap protein of PPV7. Our findings provide new insight into the prevalence of PPV7 in swine in Guangxi, China, as well as sequence data and phylogenetic analysis of these novel PPV7 isolates.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

