Single-Dose Vaccination with a Hepatotropic Adeno-associated Virus Efficiently Localizes T Cell Immunity in the Liver with the Potential To Confer Rapid Protection against Hepatitis C Virus
- PMID: 31292249
- PMCID: PMC6744243
- DOI: 10.1128/JVI.00202-19
Single-Dose Vaccination with a Hepatotropic Adeno-associated Virus Efficiently Localizes T Cell Immunity in the Liver with the Potential To Confer Rapid Protection against Hepatitis C Virus
Abstract
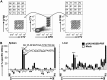
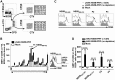
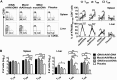
Hepatitis C virus (HCV) is a significant contributor to the global disease burden, and development of an effective vaccine is required to eliminate HCV infections worldwide. CD4+ and CD8+ T cell immunity correlates with viral clearance in primary HCV infection, and intrahepatic CD8+ tissue-resident memory T (TRM) cells provide lifelong and rapid protection against hepatotropic pathogens. Consequently, we aimed to develop a vaccine to elicit HCV-specific CD4+ and CD8+ T cells, including CD8+ TRM cells, in the liver, given that HCV primarily infects hepatocytes. To achieve this, we vaccinated wild-type BALB/c mice with a highly immunogenic cytolytic DNA vaccine encoding a model HCV (genotype 3a) nonstructural protein (NS5B) and a mutant perforin (pVAX-NS5B-PRF), as well as a recombinant adeno-associated virus (AAV) encoding NS5B (rAAV-NS5B). A novel fluorescent target array (FTA) was used to map immunodominant CD4+ T helper (TH) cell and cytotoxic CD8+ T cell epitopes of NS5B in vivo, which were subsequently used to design a KdNS5B451-459 tetramer and analyze NS5B-specific T cell responses in vaccinated mice in vivo The data showed that intradermal prime/boost vaccination with pVAX-NS5B-PRF was effective in eliciting TH and cytotoxic CD8+ T cell responses and intrahepatic CD8+ TRM cells, but a single intravenous dose of hepatotropic rAAV-NS5B was significantly more effective. As a T-cell-based vaccine against HCV should ideally result in localized T cell responses in the liver, this study describes primary observations in the context of HCV vaccination that can be used to achieve this goal.IMPORTANCE There are currently at least 71 million individuals with chronic HCV worldwide and almost two million new infections annually. Although the advent of direct-acting antivirals (DAAs) offers highly effective therapy, considerable remaining challenges argue against reliance on DAAs for HCV elimination, including high drug cost, poorly developed health infrastructure, low screening rates, and significant reinfection rates. Accordingly, development of an effective vaccine is crucial to HCV elimination. An HCV vaccine that elicits T cell immunity in the liver will be highly protective for the following reasons: (i) T cell responses against nonstructural proteins of the virus are associated with clearance of primary infection, and (ii) long-lived liver-resident T cells alone can protect against malaria infection of hepatocytes. Thus, in this study we exploit promising vaccination platforms to highlight strategies that can be used to evoke highly functional and long-lived T cell responses in the liver for protection against HCV.
Keywords: DNA vaccine; adeno-associated virus vaccine; cytotoxic T cells; helper T cells; hepatitis C virus vaccine; liver immunity.
Copyright © 2019 American Society for Microbiology.
Figures




References
-
- WHO. 2017. Global Hepatitis Report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/.
-
- WHO. 2016. Global health sector strategies for viral hepatitis 2016–2021. https://www.who.int/hepatitis/strategy2016-2021/ghss-hep/en/.
-
- Borgia SM, Hedskog C, Parhy B, Hyland RH, Stamm LM, Brainard DM, Subramanian MG, McHutchison JG, Mo H, Svarovskaia E, Shafran SD. 2018. Identification of a novel hepatitis C virus genotype from Punjab, India: expanding classification of hepatitis C virus into 8 genotypes. J Infect Dis 218:1722–1729. doi: 10.1093/infdis/jiy401. - DOI - PubMed
-
- Sacks-Davis R, Grebely J, Dore GJ, Osburn W, Cox AL, Rice TM, Spelman T, Bruneau J, Prins M, Kim AY, McGovern BH, Shoukry NH, Schinkel J, Allen TM, Morris M, Hajarizadeh B, Maher L, Lloyd AR, Page K, Hellard M, InC3 Study Group. 2015. Hepatitis C virus reinfection and spontaneous clearance of reinfection—the InC3 Study. J Infect Dis 212:1407–1419. doi: 10.1093/infdis/jiv220. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

