Emergence of ERBB2 Mutation as a Biomarker and an Actionable Target in Solid Cancers
- PMID: 31292270
- PMCID: PMC6975965
- DOI: 10.1634/theoncologist.2018-0845
Emergence of ERBB2 Mutation as a Biomarker and an Actionable Target in Solid Cancers
Abstract
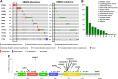
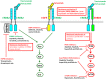
The oncogenic role ERBB2 amplification is well established in breast and gastric cancers. This has led to the development of a well-known portfolio of monoclonal antibodies and kinase inhibitors targeting the ERBB2 kinase. More recently, activating mutations in the ERBB2 gene have been increasingly reported in multiple solid cancers and were shown to play an oncogenic role similar to that of ERBB2 amplification. Thus, ERBB2 mutations define a distinct molecular subtype of solid tumors and serve as actionable targets. However, efforts to target ERBB2 mutation has met with limited clinical success, possibly because of their low frequency, inadequate understanding of the biological activity of these mutations, and difficulty in separating the drivers from the passenger mutations. Given the current impetus to deliver molecularly targeted treatments for cancer, there is an important need to understand the therapeutic potential of ERBB2 mutations. Here we review the distribution of ERBB2 mutations in different tumor types, their potential as a novel biomarker that defines new subsets in many cancers, and current data on preclinical and clinical efforts to target these mutations. IMPLICATIONS FOR PRACTICE: A current trend in oncology is to identify novel genomic drivers of solid tumors and developing precision treatments that target them. ERBB2 amplification is an established therapeutic target in breast and gastric cancers, but efforts to translate this finding to other solid tumors with ERBB2 amplification have not been effective. Recently the focus has turned to targeting activating ERBB2 mutations. The year 2018 marked an important milestone in establishing ERBB2 mutation as an important actionable target in multiple cancer types. There have been several recent preclinical and clinical studies evaluating ERBB2 mutation as a therapeutic target with varying success. With increasing access to next-generation sequencing technologies in the clinic, oncologists are frequently identifying activating ERBB2 mutations in patients with cancer. There is a significant need both from the clinician and bench scientist perspectives to understand the current state of affairs for ERBB2 mutations.
Keywords: ERBB2 mutation; Gastrointestinal cancer; HER2; Non‐small cell lung cancer; Tyrosine kinase.
© AlphaMed Press 2019.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures


References
-
- Yarden Y and Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

