Integrative multi-omic analysis identifies new drivers and pathways in molecularly distinct subtypes of ALS
- PMID: 31292500
- PMCID: PMC6620285
- DOI: 10.1038/s41598-019-46355-w
Integrative multi-omic analysis identifies new drivers and pathways in molecularly distinct subtypes of ALS
Abstract
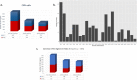
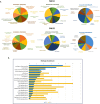
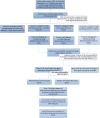
Amyotrophic lateral sclerosis (ALS) is an incurable and fatal neurodegenerative disease. Increasing the chances of success for future clinical strategies requires more in-depth knowledge of the molecular basis underlying disease heterogeneity. We recently laid the foundation for a molecular taxonomy of ALS by whole-genome expression profiling of motor cortex from sporadic ALS (SALS) patients. Here, we analyzed copy number variants (CNVs) occurring in the same patients, by using a customized exon-centered comparative genomic hybridization array (aCGH) covering a large panel of ALS-related genes. A large number of novel and known disease-associated CNVs were detected in SALS samples, including several subgroup-specific loci, suggestive of a great divergence of two subgroups at the molecular level. Integrative analysis of copy number profiles with their associated transcriptomic data revealed subtype-specific genomic perturbations and candidate driver genes positively correlated with transcriptional signatures, suggesting a strong interaction between genomic and transcriptomic events in ALS pathogenesis. The functional analysis confirmed our previous pathway-based characterization of SALS subtypes and identified 24 potential candidates for genomic-based patient stratification. To our knowledge, this is the first comprehensive "omics" analysis of molecular events characterizing SALS pathology, providing a road map to facilitate genome-guided personalized diagnosis and treatments for this devastating disease.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

