Generation and characterization of a stable cell line persistently replicating and secreting the human hepatitis delta virus
- PMID: 31292511
- PMCID: PMC6620269
- DOI: 10.1038/s41598-019-46493-1
Generation and characterization of a stable cell line persistently replicating and secreting the human hepatitis delta virus
Abstract
Human hepatitis delta virus (HDV) causes the most severe form of viral hepatitis. Approximately 15-25 million people are chronically infected with HDV. As a satellite virus of the human hepatitis B virus (HBV), HDV uses the HBV-encoded envelope proteins for egress from and de novo entry into hepatocytes. So far, in vitro production of HDV particles is restricted to co-transfection of cells with HDV/HBV encoding cDNAs. This approach has several limitations. In this study, we established HuH7-END cells, which continuously secrete infectious HDV virions. The cell line was generated through stepwise stable integration of the cDNA of the HDV antigenome, the genes for the HBV envelope proteins and the HBV/HDV receptor NTCP. We found that HuH7-END cells release infectious HDV particles up to 400 million copies/milliliter and support virus spread to co-cultured cells. Due to the expression of NTCP, HuH7-END cells are also susceptible to de novo HDV entry. Virus production is stable for >16 passages and can be scaled up for preparation of large HDV virus stocks. Finally, HuH7-END cells are suitable for screening of antiviral drugs targeting HDV replication. In summary, the HuH7-END cell line provides a novel tool to study HDV replication in vitro.
Conflict of interest statement
Stephan Urban is co-applicant and co-inventor on patents protecting HBV preS-derived lipopeptides (Myrcludex B) for the use of HBV/HDV entry inhibitors. The other authors in this study declare nothing to disclose.
Figures
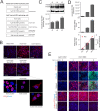
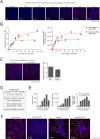
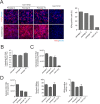
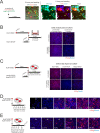

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

