Structural analysis of the Aβ(11-42) amyloid fibril based on hydrophobicity distribution
- PMID: 31292794
- PMCID: PMC6687686
- DOI: 10.1007/s10822-019-00209-9
Structural analysis of the Aβ(11-42) amyloid fibril based on hydrophobicity distribution
Abstract
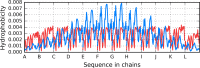
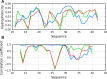
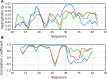
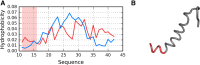
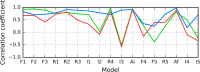
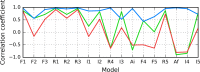
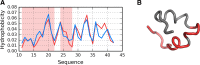
The structure of the Aβ(11-42) amyloid available in PDB makes possible the molecular analysis of the specificity of amyloid formation. This molecule (PDB ID 2MVX) is the object of analysis. This work presents the outcome of in silico experiments involving various alternative conformations of the Aβ(11-42) sequence, providing clues as to the amylodogenecity of its constituent fragments. The reference structure (PDB) has been compared with folds generated using I-Tasser and Robetta-the strongest contenders in the CASP challenge. Additionally, a polypeptide which matches the Aβ(11-42) sequence has been subjected to folding simulations based on the fuzzy oil drop model, which favors the production of a monocentric hydrophobic core. Computer simulations yielded 15 distinct structural forma (five per software package), which, when compared to the experimentally determined structure, allow us to study the role of structural elements which cause an otherwise globular protein to transform into an amyloid. The unusual positions of hydrophilic residues disrupting the expected hydrophobic core and propagating linearly along the long axis of fibril is recognized as the seed for amyloidogenic transformation in this polypeptide. This paper discusses the structure of the Aβ(11-42) amyloid fibril, listed in PDB under ID 2MXU (fragment od Aβ(1-42) amyloid).
Keywords: Amyloidosis; Aβ(11–42); Aβ(1–42); Fibrillation.
Figures


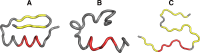
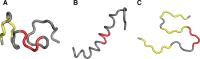

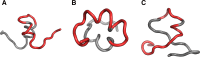

References
-
- Khoury GA, Liwo A, Khatib F, Zhou H, Chopra G, Bacardit J, Bortot LO, Faccioli RA, Deng X, He Y, Krupa P, Li J, Mozolewska MA, Sieradzan AK, Smadbeck J, Wirecki T, Cooper S, Flatten J, Xu K, Baker D, Cheng J, Delbem AC, Floudas CA, Keasar C, Levitt M, Popović Z, Scheraga HA, Skolnick J, Crivelli SN, Foldit Players WeFold: a coopetition for protein structure prediction. Proteins. 2014;82(9):1850–1868. doi: 10.1002/prot.24538. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

