Not Just a Pathogen? Description of a Plant-Beneficial Pseudomonas syringae Strain
- PMID: 31293547
- PMCID: PMC6598456
- DOI: 10.3389/fmicb.2019.01409
Not Just a Pathogen? Description of a Plant-Beneficial Pseudomonas syringae Strain
Abstract
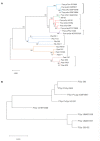
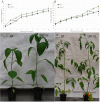
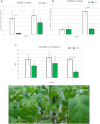
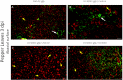
Plants develop in a microbe-rich environment and must interact with a plethora of microorganisms, both pathogenic and beneficial. Indeed, such is the case of Pseudomonas, and its model organisms P. fluorescens and P. syringae, a bacterial genus that has received particular attention because of its beneficial effect on plants and its pathogenic strains. The present study aims to compare plant-beneficial and pathogenic strains belonging to the P. syringae species to get new insights into the distinction between the two types of plant-microbe interactions. In assays carried out under greenhouse conditions, P. syringae pv. syringae strain 260-02 was shown to promote plant-growth and to exert biocontrol of P. syringae pv. tomato strain DC3000, against the Botrytis cinerea fungus and the Cymbidium Ringspot Virus. This P. syringae strain also had a distinct volatile emission profile, as well as a different plant-colonization pattern, visualized by confocal microscopy and gfp labeled strains, compared to strain DC3000. Despite the different behavior, the P. syringae strain 260-02 showed great similarity to pathogenic strains at a genomic level. However, genome analyses highlighted a few differences that form the basis for the following hypotheses regarding strain 260-02. P. syringae strain 260-02: (i) possesses non-functional virulence genes, like the mangotoxin-producing operon Mbo; (ii) has different regulation pathways, suggested by the difference in the autoinducer system and the lack of a virulence activator gene; (iii) has genes encoding DNA methylases different from those found in other P. syringae strains, suggested by the presence of horizontal-gene-transfer-obtained methylases that could affect gene expression.
Keywords: Botrytis cinerea; Pseudomonas syringae; biocontrol; confocal microscopy; pangenome analysis.
Figures




References
-
- Antunes L. C. M., Ferreira R. B. R. (2009). Intercellular communication in bacteria. Crit. Rev. Microbiol. 3 69–80. - PubMed
-
- Arrebola E., Cazorla F. M., Codina J. C., Gutierrez-Barranquero J. A., Perez-Garcia A., de Vicente A. (2009). Contribution of mangotoxin to the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Int. Microbiol. 12 87–95. - PubMed

