Genomic analyses suggest adaptive differentiation of northern European native cattle breeds
- PMID: 31293626
- PMCID: PMC6597895
- DOI: 10.1111/eva.12783
Genomic analyses suggest adaptive differentiation of northern European native cattle breeds
Abstract
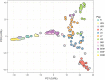
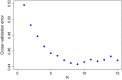
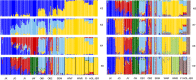
Native domestic breeds represent important cultural heritage and genetic diversity relevant for production traits, environmental adaptation and food security. However, risks associated with low effective population size, such as inbreeding and genetic drift, have elevated concerns over whether unique within-breed lineages should be kept separate or managed as one population. As a conservation genomic case study of the genetic diversity represented by native breeds, we examined native and commercial cattle (Bos taurus) breeds including the threatened Danish Jutland cattle. We examined population structure and genetic diversity within breeds and lineages genotyped across 770K single nucleotide polymorphism loci to determine (a) the amount and distribution of genetic diversity in native breeds, and (b) the role of genetic drift versus selection. We further investigated the presence of outlier loci to detect (c) signatures of environmental selection in native versus commercial breeds, and (d) native breed adaptation to various landscapes. Moreover, we included older cryopreserved samples to determine (e) whether cryopreservation allows (re)introduction of original genetic diversity. We investigated a final set of 195 individuals and 677K autosomal loci for genetic diversity within and among breeds, examined population structure with principal component analyses and a maximum-likelihood approach and searched for outlier loci suggesting artificial or natural selection. Our findings demonstrate the potential of genomics for identifying the uniqueness of native domestic breeds, and for maintaining their genetic diversity and long-term evolutionary potential through conservation plans balancing inbreeding with carefully designed outcrossing. One promising opportunity is the use of cryopreserved samples, which can provide important genetic diversity for populations with few individuals, while helping to preserve their traditional genetic characteristics. Outlier tests for native versus commercial breeds identified genes associated with climate adaptation, immunity and metabolism, and native breeds may carry genetic variation important for animal health and robustness in a changing climate.
Keywords: Bos taurus; animal health; artificial selection; climate adaptation; conservation genomics; environmental selection; production traits; single nucleotide polymorphism.
Conflict of interest statement
None declared.
Figures

References
-
- Alexander, D. H. , Shringarpure, S. S. , Novembre, J. , & Lange, K. (2015). Admixture 1.3 software manual. Retrieved from https://www.genetics.ucla.edu/software/admixture/admixture-manual.pdf
-
- Bailey, D. W. , Lunt, S. , Lipka, A. , Thomas, M. G. , Medrano, J. F. , Cánovas, A. , … Jensen, D. (2015). Genetic influences on cattle grazing distribution: Association of genetic markers with terrain use in cattle. Rangeland Ecology Management, 68, 142–149. 10.1016/j.rama.2015.02.001 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

