CASP13 target classification into tertiary structure prediction categories
- PMID: 31294862
- PMCID: PMC6851465
- DOI: 10.1002/prot.25775
CASP13 target classification into tertiary structure prediction categories
Abstract
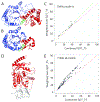
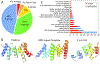
Protein target structures for the Critical Assessment of Structure Prediction round 13 (CASP13) were split into evaluation units (EUs) based on their structural domains, the domain organization of available templates, and the performance of servers on whole targets compared to split target domains. Eighty targets were split into 112 EUs. The EUs were classified into categories suitable for assessment of high accuracy modeling (or template-based modeling [TBM]) and topology (or free modeling [FM]) based on target difficulty. Assignment into assessment categories considered the following criteria: (a) the evolutionary relationship of target domains to existing fold space as defined by the Evolutionary Classification of Protein Domains (ECOD) database; (b) the clustering of target domains using eight objective sequence, structure, and performance measures; and (c) the placement of target domains in a scatter plot of target difficulty against server performance used in the previous CASP. Generally, target domains with good server predictions had close template homologs and were classified as TBM. Alternately, targets with poor server predictions represent a mixture of fast evolving homologs, structure analogs, and new folds, and were classified as FM or FM/TBM overlap.
Keywords: CASP13; classification; fold space; free modeling; protein structure; sequence homologs; structure analogs; structure prediction; template-based modeling.
© 2019 Wiley Periodicals, Inc.
Figures