A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth
- PMID: 31295257
- PMCID: PMC6650080
- DOI: 10.1371/journal.pbio.3000085
A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth
Abstract
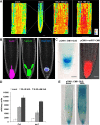
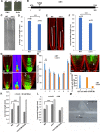
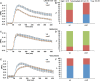
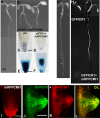
Signaling cross talks between auxin, a regulator of plant development, and Ca2+, a universal second messenger, have been proposed to modulate developmental plasticity in plants. However, the underlying molecular mechanisms are largely unknown. Here, we report that in Arabidopsis roots, auxin elicits specific Ca2+ signaling patterns that spatially coincide with the expression pattern of auxin-regulated genes. We have identified the single EF-hand Ca2+-binding protein Ca2+-dependent modulator of ICR1 (CMI1) as an interactor of the Rho of plants (ROP) effector interactor of constitutively active ROP (ICR1). CMI1 expression is directly up-regulated by auxin, whereas the loss of function of CMI1 associates with the repression of auxin-induced Ca2+ increases in the lateral root cap and vasculature, indicating that CMI1 represses early auxin responses. In agreement, cmi1 mutants display an increased auxin response including shorter primary roots, longer root hairs, longer hypocotyls, and altered lateral root formation. Binding to ICR1 affects subcellular localization of CMI1 and its function. The interaction between CMI1 and ICR1 is Ca2+-dependent and involves a conserved hydrophobic pocket in CMI1 and calmodulin binding-like domain in ICR1. Remarkably, CMI1 is monomeric in solution and in vitro changes its secondary structure at cellular resting Ca2+ concentrations ranging between 10-9 and 10-8 M. Hence, CMI1 is a Ca2+-dependent transducer of auxin-regulated gene expression, which can function in a cell-specific fashion at steady-state as well as at elevated cellular Ca2+ levels to regulate auxin responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

